| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Voltage-dependent N-type calcium channel subunit alpha-1B |
|---|
| Ligand | BDBM61724 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Homogeneous Time-Resolved Fluorescence Resonance Energy Transfer (HTRF) Assay |
|---|
| IC50 | 1840±n/a nM |
|---|
| Citation |  PubChem, PC Homogeneous Time-Resolved Fluorescence Resonance Energy Transfer (HTRF) Assay PubChem Bioassay(2009)[AID] PubChem, PC Homogeneous Time-Resolved Fluorescence Resonance Energy Transfer (HTRF) Assay PubChem Bioassay(2009)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Voltage-dependent N-type calcium channel subunit alpha-1B |
|---|
| Name: | Voltage-dependent N-type calcium channel subunit alpha-1B |
|---|
| Synonyms: | BIII | Brain calcium channel III | CAC1B_HUMAN | CACH5 | CACNA1B | CACNL1A5 | Calcium channel (Type N) | Calcium channel, L type, alpha-1 polypeptide isoform 5 | Voltage-dependent N-type calcium channel subunit alpha-1B | Voltage-dependent N-type calcium channel subunit alpha-1B/Voltage-dependent calcium channel subunit alpha-2/delta-1/Voltage-dependent L-type calcium channel subunit beta-3 | Voltage-gated N-type calcium channel alpha-1B subunit | Voltage-gated N-type calcium channel alpha-1B subunit/Amyloid beta A4 precursor protein-binding family A member 1 | Voltage-gated calcium channel | Voltage-gated calcium channel subunit alpha Cav2.2 | Voltage-gated calcium channel subunit alpha Cav2.2 ((alpha 1B, beta 1b, alpha 2 delta-1) | calcium channel, voltage-dependent, N type, alpha 1B subunit |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 262548.16 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q00975 |
|---|
| Residue: | 2339 |
|---|
| Sequence: | MVRFGDELGGRYGGPGGGERARGGGAGGAGGPGPGGLQPGQRVLYKQSIAQRARTMALYN
PIPVKQNCFTVNRSLFVFSEDNVVRKYAKRITEWPPFEYMILATIIANCIVLALEQHLPD
GDKTPMSERLDDTEPYFIGIFCFEAGIKIIALGFVFHKGSYLRNGWNVMDFVVVLTGILA
TAGTDFDLRTLRAVRVLRPLKLVSGIPSLQVVLKSIMKAMVPLLQIGLLLFFAILMFAII
GLEFYMGKFHKACFPNSTDAEPVGDFPCGKEAPARLCEGDTECREYWPGPNFGITNFDNI
LFAILTVFQCITMEGWTDILYNTNDAAGNTWNWLYFIPLIIIGSFFMLNLVLGVLSGEFA
KERERVENRRAFLKLRRQQQIERELNGYLEWIFKAEEVMLAEEDRNAEEKSPLDVLKRAA
TKKSRNDLIHAEEGEDRFADLCAVGSPFARASLKSGKTESSSYFRRKEKMFRFFIRRMVK
AQSFYWVVLCVVALNTLCVAMVHYNQPRRLTTTLYFAEFVFLGLFLTEMSLKMYGLGPRS
YFRSSFNCFDFGVIVGSVFEVVWAAIKPGSSFGISVLRALRLLRIFKVTKYWSSLRNLVV
SLLNSMKSIISLLFLLFLFIVVFALLGMQLFGGQFNFQDETPTTNFDTFPAAILTVFQIL
TGEDWNAVMYHGIESQGGVSKGMFSSFYFIVLTLFGNYTLLNVFLAIAVDNLANAQELTK
DEEEMEEAANQKLALQKAKEVAEVSPMSAANISIAARQQNSAKARSVWEQRASQLRLQNL
RASCEALYSEMDPEERLRFATTRHLRPDMKTHLDRPLVVELGRDGARGPVGGKARPEAAE
APEGVDPPRRHHRHRDKDKTPAAGDQDRAEAPKAESGEPGAREERPRPHRSHSKEAAGPP
EARSERGRGPGPEGGRRHHRRGSPEEAAEREPRRHRAHRHQDPSKECAGAKGERRARHRG
GPRAGPREAESGEEPARRHRARHKAQPAHEAVEKETTEKEATEKEAEIVEADKEKELRNH
QPREPHCDLETSGTVTVGPMHTLPSTCLQKVEEQPEDADNQRNVTRMGSQPPDPNTIVHI
PVMLTGPLGEATVVPSGNVDLESQAEGKKEVEADDVMRSGPRPIVPYSSMFCLSPTNLLR
RFCHYIVTMRYFEVVILVVIALSSIALAAEDPVRTDSPRNNALKYLDYIFTGVFTFEMVI
KMIDLGLLLHPGAYFRDLWNILDFIVVSGALVAFAFSGSKGKDINTIKSLRVLRVLRPLK
TIKRLPKLKAVFDCVVNSLKNVLNILIVYMLFMFIFAVIAVQLFKGKFFYCTDESKELER
DCRGQYLDYEKEEVEAQPRQWKKYDFHYDNVLWALLTLFTVSTGEGWPMVLKHSVDATYE
EQGPSPGYRMELSIFYVVYFVVFPFFFVNIFVALIIITFQEQGDKVMSECSLEKNERACI
DFAISAKPLTRYMPQNRQSFQYKTWTFVVSPPFEYFIMAMIALNTVVLMMKFYDAPYEYE
LMLKCLNIVFTSMFSMECVLKIIAFGVLNYFRDAWNVFDFVTVLGSITDILVTEIAETNN
FINLSFLRLFRAARLIKLLRQGYTIRILLWTFVQSFKALPYVCLLIAMLFFIYAIIGMQV
FGNIALDDDTSINRHNNFRTFLQALMLLFRSATGEAWHEIMLSCLSNQACDEQANATECG
SDFAYFYFVSFIFLCSFLMLNLFVAVIMDNFEYLTRDSSILGPHHLDEFIRVWAEYDPAA
CGRISYNDMFEMLKHMSPPLGLGKKCPARVAYKRLVRMNMPISNEDMTVHFTSTLMALIR
TALEIKLAPAGTKQHQCDAELRKEISVVWANLPQKTLDLLVPPHKPDEMTVGKVYAALMI
FDFYKQNKTTRDQMQQAPGGLSQMGPVSLFHPLKATLEQTQPAVLRGARVFLRQKSSTSL
SNGGAIQNQESGIKESVSWGTQRTQDAPHEARPPLERGHSTEIPVGRSGALAVDVQMQSI
TRRGPDGEPQPGLESQGRAASMPRLAAETQPVTDASPMKRSISTLAQRPRGTHLCSTTPD
RPPPSQASSHHHHHRCHRRRDRKQRSLEKGPSLSADMDGAPSSAVGPGLPPGEGPTGCRR
ERERRQERGRSQERRQPSSSSSEKQRFYSCDRFGGREPPKPKPSLSSHPTSPTAGQEPGP
HPQGSGSVNGSPLLSTSGASTPGRGGRRQLPQTPLTPRPSITYKTANSSPIHFAGAQTSL
PAFSPGRLSRGLSEHNALLQRDPLSQPLAPGSRIGSDPYLGQRLDSEASVHALPEDTLTF
EEAVATNSGRSSRTSYVSSLTSQSHPLRRVPNGYHCTLGLSSGGRARHSYHHPDQDHWC
|
|
|
|---|
| BDBM61724 |
|---|
| n/a |
|---|
| Name | BDBM61724 |
|---|
| Synonyms: | (8S)-3-(3-methacrylamidophenyl)-7-(1-methylcyclopropanecarbonyl)-1-oxa-2,7-diazaspiro[4.4]non-2-ene-8-carboxamide | (8S)-7-(1-methylcyclopropanecarbonyl)-3-[3-(2-methylprop-2-enoylamino)phenyl]-1-oxa-2,7-diazaspiro[4.4]non-2-ene-8-carboxamide | (8S)-7-(1-methylcyclopropyl)carbonyl-3-[3-(2-methylprop-2-enoylamino)phenyl]-1-oxa-2,7-diazaspiro[4.4]non-2-ene-8-carboxamide | (8S)-7-[(1-methylcyclopropyl)-oxomethyl]-3-[3-[(2-methyl-1-oxoprop-2-enyl)amino]phenyl]-1-oxa-2,7-diazaspiro[4.4]non-2-ene-8-carboxamide | MLS000562014 | SMR000390649 | cid_16745487 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H26N4O4 |
|---|
| Mol. Mass. | 410.4662 |
|---|
| SMILES | CC(=C)C(=O)Nc1cccc(c1)C1=NOC2(C[C@H](N(C2)C(=O)C2(C)CC2)C(N)=O)C1 |t:13| |
|---|
| Structure | 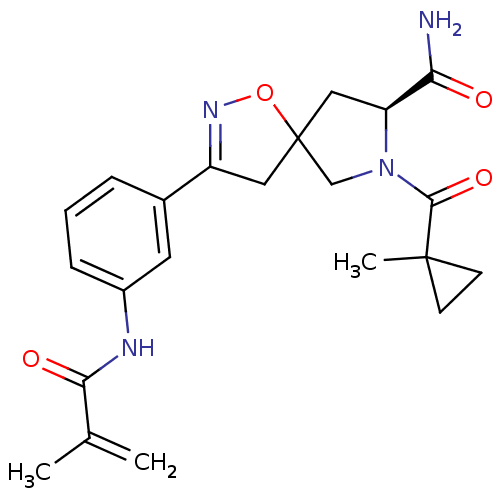
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Homogeneous Time-Resolved Fluorescence Resonance Energy Transfer (HTRF) Assay PubChem Bioassay(2009)[AID]
PubChem, PC Homogeneous Time-Resolved Fluorescence Resonance Energy Transfer (HTRF) Assay PubChem Bioassay(2009)[AID]