| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Epstein-Barr nuclear antigen 1 |
|---|
| Ligand | BDBM50995 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of the Epstein-Barr virus nuclear antigen 1 (EBNA-1) |
|---|
| IC50 | 14533±n/a nM |
|---|
| Citation |  PubChem, PC Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of the Epstein-Barr virus nuclear antigen 1 (EBNA-1) PubChem Bioassay(2010)[AID] PubChem, PC Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of the Epstein-Barr virus nuclear antigen 1 (EBNA-1) PubChem Bioassay(2010)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Epstein-Barr nuclear antigen 1 |
|---|
| Name: | Epstein-Barr nuclear antigen 1 |
|---|
| Synonyms: | EBNA-1 protein | EBNA1 | EBNA1_EBVB9 | Epstein-Barr virus protease (EBV Pr) |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 56444.81 |
|---|
| Organism: | Human herpesvirus 4 |
|---|
| Description: | gi_23893623 |
|---|
| Residue: | 641 |
|---|
| Sequence: | MSDEGPGTGPGNGLGEKGDTSGPEGSGGSGPQRRGGDNHGRGRGRGRGRGGGRPGAPGGS
GSGPRHRDGVRRPQKRPSCIGCKGTHGGTGAGAGAGGAGAGGAGAGGGAGAGGGAGGAGG
AGGAGAGGGAGAGGGAGGAGGAGAGGGAGAGGGAGGAGAGGGAGGAGGAGAGGGAGAGGG
AGGAGAGGGAGGAGGAGAGGGAGAGGAGGAGGAGAGGAGAGGGAGGAGGAGAGGAGAGGA
GAGGAGAGGAGGAGAGGAGGAGAGGAGGAGAGGGAGGAGAGGGAGGAGAGGAGGAGAGGA
GGAGAGGAGGAGAGGGAGAGGAGAGGGGRGRGGSGGRGRGGSGGRGRGGSGGRRGRGRER
ARGGSRERARGRGRGRGEKRPRSPSSQSSSSGSPPRRPPPGRRPFFHPVGEADYFEYHQE
GGPDGEPDVPPGAIEQGPADDPGEGPSTGPRGQGDGGRRKKGGWFGKHRGQGGSNPKFEN
IAEGLRALLARSHVERTTDEGTWVAGVFVYGGSKTSLYNLRRGTALAIPQCRLTPLSRLP
FGMAPGPGPQPGPLRESIVCYFMVFLQTHIFAEVLKDAIKDLVMTKPAPTCNIRVTVCSF
DDGVDLPPWFPPMVEGAAAEGDDGDDGDEGGDGDEGEEGQE
|
|
|
|---|
| BDBM50995 |
|---|
| n/a |
|---|
| Name | BDBM50995 |
|---|
| Synonyms: | 2-{4-[(4-Hydroxy-benzoyl)-hydrazonomethyl]-phenoxy}-N-(4-nitro-phenyl)-acetamide | 4-hydroxy-N-[[4-[2-(4-nitroanilino)-2-oxoethoxy]phenyl]methylideneamino]benzamide | 4-hydroxy-N-[[4-[2-keto-2-(4-nitroanilino)ethoxy]benzylidene]amino]benzamide | MLS000548295 | N-[[4-[2-[(4-nitrophenyl)amino]-2-oxidanylidene-ethoxy]phenyl]methylideneamino]-4-oxidanyl-benzamide | SMR000171643 | cid_3115893 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H18N4O6 |
|---|
| Mol. Mass. | 434.4015 |
|---|
| SMILES | Oc1ccc(cc1)C(=O)NN=Cc1ccc(OCC(=O)Nc2ccc(cc2)[N+]([O-])=O)cc1 |w:10.10| |
|---|
| Structure | 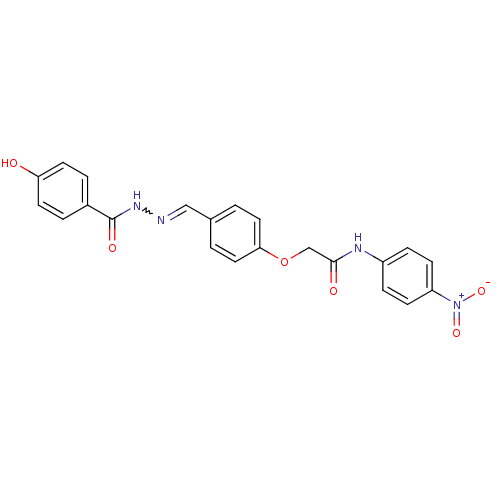
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of the Epstein-Barr virus nuclear antigen 1 (EBNA-1) PubChem Bioassay(2010)[AID]
PubChem, PC Fluorescence polarization-based biochemical high throughput dose response assay for inhibitors of the Epstein-Barr virus nuclear antigen 1 (EBNA-1) PubChem Bioassay(2010)[AID]