| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Heat shock factor protein 1 |
|---|
| Ligand | BDBM62726 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Luminescence Cell-Based Dose Confimation HTS to Identify Inhibitors of Heat Shock Factor 1 (HSF1) |
|---|
| EC50 | 69891±1738 nM |
|---|
| Citation |  PubChem, PC Luminescence Cell-Based Dose Confimation HTS to Identify Inhibitors of Heat Shock Factor 1 (HSF1) PubChem Bioassay(2010)[AID] PubChem, PC Luminescence Cell-Based Dose Confimation HTS to Identify Inhibitors of Heat Shock Factor 1 (HSF1) PubChem Bioassay(2010)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Heat shock factor protein 1 |
|---|
| Name: | Heat shock factor protein 1 |
|---|
| Synonyms: | HSF1_MOUSE | Hsf1 | Hsf1 protein |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 57208.68 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | P38532 |
|---|
| Residue: | 525 |
|---|
| Sequence: | MDLAVGPGAAGPSNVPAFLTKLWTLVSDPDTDALICWSPSGNSFHVFDQGQFAKEVLPKY
FKHNNMASFVRQLNMYGFRKVVHIEQGGLVKPERDDTEFQHPCFLRGQEQLLENIKRKVT
SVSTLKSEDIKIRQDSVTRLLTDVQLMKGKQECMDSKLLAMKHENEALWREVASLRQKHA
QQQKVVNKLIQFLISLVQSNRILGVKRKIPLMLSDSNSAHSVPKYGRQYSLEHVHGPGPY
SAPSPAYSSSSLYSSDAVTSSGPIISDITELAPTSPLASPGRSIDERPLSSSTLVRVKQE
PPSPPHSPRVLEASPGRPSSMDTPLSPTAFIDSILRESEPTPAASNTAPMDTTGAQAPAL
PTPSTPEKCLSVACLDKNELSDHLDAMDSNLDNLQTMLTSHGFSVDTSALLDLFSPSVTM
PDMSLPDLDSSLASIQELLSPQEPPRPIEAENSNPDSGKQLVHYTAQPLFLLDPDAVDTG
SSELPVLFELGESSYFSEGDDYTDDPTISLLTGTEPHKAKDPTVS
|
|
|
|---|
| BDBM62726 |
|---|
| n/a |
|---|
| Name | BDBM62726 |
|---|
| Synonyms: | (4Z)-5-(2-furanyl)-4-[hydroxy-(2-methyl-2,3-dihydrobenzofuran-5-yl)methylidene]-1-(2-oxolanylmethyl)pyrrolidine-2,3-dione | (4Z)-5-(2-furyl)-4-[hydroxy-(2-methylcoumaran-5-yl)methylene]-1-(tetrahydrofurfuryl)pyrrolidine-2,3-quinone | (4Z)-5-(furan-2-yl)-4-[(2-methyl-2,3-dihydro-1-benzofuran-5-yl)-oxidanyl-methylidene]-1-(oxolan-2-ylmethyl)pyrrolidine-2,3-dione | (4Z)-5-(furan-2-yl)-4-[hydroxy-(2-methyl-2,3-dihydro-1-benzofuran-5-yl)methylidene]-1-(oxolan-2-ylmethyl)pyrrolidine-2,3-dione | 5-(2-furyl)-3-hydroxy-4-[(2-methyl-2,3-dihydro-1-benzofuran-5-yl)carbonyl]-1-(tetrahydro-2-furanylmethyl)-1,5-dihydro-2H-pyrrol-2-one | MLS000049070 | SMR000074348 | cid_5771719 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H23NO6 |
|---|
| Mol. Mass. | 409.4318 |
|---|
| SMILES | CC1Cc2cc(ccc2O1)C(=O)C1C(N(CC2CCCO2)C(=O)C1=O)c1ccco1 |
|---|
| Structure | 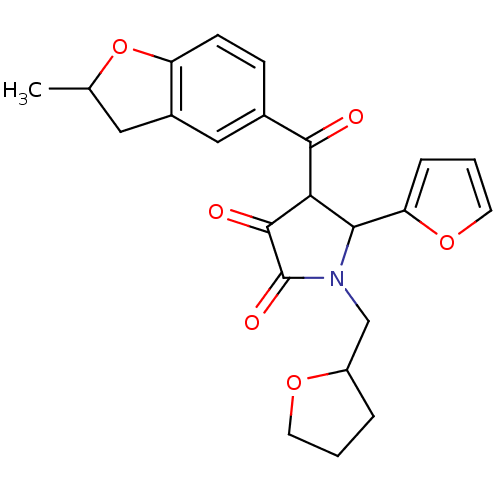
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Luminescence Cell-Based Dose Confimation HTS to Identify Inhibitors of Heat Shock Factor 1 (HSF1) PubChem Bioassay(2010)[AID]
PubChem, PC Luminescence Cell-Based Dose Confimation HTS to Identify Inhibitors of Heat Shock Factor 1 (HSF1) PubChem Bioassay(2010)[AID]