| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-dependent molecular chaperone HSP82 |
|---|
| Ligand | BDBM36888 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Luminesence Cell-Based Secondary Assay to Identify Inhibitors of Hsp90 |
|---|
| EC50 | >150000±0 nM |
|---|
| Citation |  PubChem, PC Luminesence Cell-Based Secondary Assay to Identify Inhibitors of Hsp90 PubChem Bioassay(2010)[AID] PubChem, PC Luminesence Cell-Based Secondary Assay to Identify Inhibitors of Hsp90 PubChem Bioassay(2010)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| ATP-dependent molecular chaperone HSP82 |
|---|
| Name: | ATP-dependent molecular chaperone HSP82 |
|---|
| Synonyms: | heat shock protein 90 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 80784.12 |
|---|
| Organism: | Candida albicans |
|---|
| Description: | gi_994798 |
|---|
| Residue: | 707 |
|---|
| Sequence: | MADAKVETHEFTAEISQLMSLIINTVYSNKEIFLRELISNASDALDKIRYQALSDPSQLE
SEPELFIRIIPQKDQKVLEIRDSGIGMTKADLVNNLGTIAKSGTKSFMEALSAGADVSMI
GQFGVGFYSLFLVADHVQVISKHNDDEQYVWESNAGGKFTVTLDETNERLGRGTMLRLFL
KEDQLEYLEEKRIKEVVKKHSEFVAYPIQLVVTKEVEKEVPETEEEDKAAEEDDKKPKLE
EVKDEEDEKKEKKTKTVKEEVTETEELNKTKPLWTRNPSDITQDEYNAFYKSISNDWEDP
LAVKHFSVEGQLEFRAILFVPKRAPFDAFESKKKKNNIKLYVRRVFITDDAEELIPEWLS
FIKGVVDSEDLPLNLSREMLQQNKILKVIRKNIVKKMIETFNEISEDQEQFNQFYTAFSK
NIKLGIHEDAQNRQSLAKLLRFYSTKSSEEMTSLSDYVTRMPEHQKNIYYITGESIKAVE
KSPFLDALKAKNFEVLFMVDPIDEYAMTQLKEFEDKKLVDITKDFELEESDEEKAAREKE
IKEYEPLTKALKDILGDQVEKVVVSYKLVDAPAAIRTGQFGWSANMERIMKAQALRDTTM
SSYMSSKKTFEISPSSPIIKELKKKVETDGAEDKTVKDLTTLLFDTALLTSGFTLDEPSN
FAHRINRLIALGLNIDDDSEETAVEPEATTTASTDEPAGESAMEEVD
|
|
|
|---|
| BDBM36888 |
|---|
| n/a |
|---|
| Name | BDBM36888 |
|---|
| Synonyms: | 4-(3,4-dihydro-1H-isoquinolin-2-yl)-5H-pyrimid[5,4-b]indole | 4-(3,4-dihydro-1H-isoquinolin-2-yl)-5H-pyrimido[5,4-b]indole | MLS000078381 | SMR000039631 | cid_658734 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H16N4 |
|---|
| Mol. Mass. | 300.3571 |
|---|
| SMILES | C1Cc2ccccc2CN1c1ncnc2c3ccccc3[nH]c12 |
|---|
| Structure | 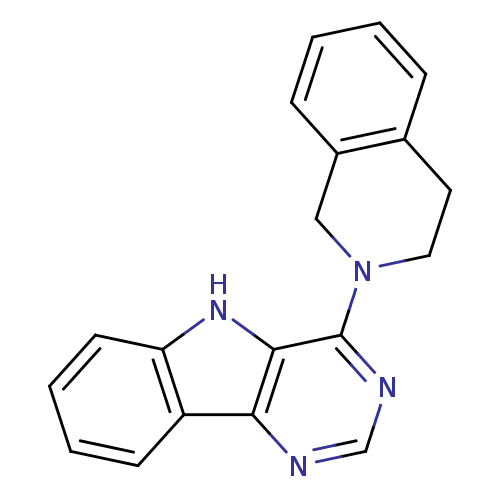
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Luminesence Cell-Based Secondary Assay to Identify Inhibitors of Hsp90 PubChem Bioassay(2010)[AID]
PubChem, PC Luminesence Cell-Based Secondary Assay to Identify Inhibitors of Hsp90 PubChem Bioassay(2010)[AID]