

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Ion channel NompC | ||
| Ligand | BDBM46991 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Late stage counterscreen for the probe development effort to identify selective agonists of the Transient Receptor Potential Channels 3 (TRPML3) | ||
| EC50 | >29900±n/a nM | ||
| Citation |  PubChem, PC Late stage counterscreen for the probe development effort to identify selective agonists of the Transient Receptor Potential Channels 3 (TRPML3): fluorescence-based cell-based dose response assay for TRPN1 agonists. PubChem Bioassay(2010)[AID] PubChem, PC Late stage counterscreen for the probe development effort to identify selective agonists of the Transient Receptor Potential Channels 3 (TRPML3): fluorescence-based cell-based dose response assay for TRPN1 agonists. PubChem Bioassay(2010)[AID] | ||
| More Info.: | Get all data from this article, Solution Info, Assay Method | ||
| Ion channel NompC | |||
| Name: | Ion channel NompC | ||
| Synonyms: | transient receptor potential cation channel, subfamily N, member 1 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 175824.78 | ||
| Organism: | Danio rerio | ||
| Description: | Q7T1G6 | ||
| Residue: | 1614 | ||
| Sequence: |
| ||
| BDBM46991 | |||
| n/a | |||
| Name | BDBM46991 | ||
| Synonyms: | 3-(4-chlorophenyl)-5-methyl-4-(2-tosylpyrazol-3-yl)isoxazole | 3-(4-chlorophenyl)-5-methyl-4-[2-(4-methylphenyl)sulfonyl-3-pyrazolyl]isoxazole | 3-(4-chlorophenyl)-5-methyl-4-[2-(4-methylphenyl)sulfonylpyrazol-3-yl]-1,2-oxazole | 3-(4-chlorophenyl)-5-methyl-4-{1-[(4-methylphenyl)sulfonyl]-1H-pyrazol-5-yl}isoxazole | MLS001111122 | SMR000457056 | cid_2745583 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H16ClN3O3S | ||
| Mol. Mass. | 413.877 | ||
| SMILES | Cc1onc(c1-c1ccnn1S(=O)(=O)c1ccc(C)cc1)-c1ccc(Cl)cc1 |(6.9,-2.18,;7.81,-.93,;9.35,-.93,;9.82,.53,;8.58,1.44,;7.33,.53,;5.87,1.01,;5.39,2.47,;3.85,2.47,;3.37,1.01,;4.62,.1,;4.62,-1.44,;6.16,-1.44,;3.08,-1.44,;4.62,-2.98,;3.29,-3.75,;3.29,-5.29,;4.62,-6.06,;4.62,-7.6,;5.95,-5.29,;5.95,-3.75,;8.58,2.98,;7.24,3.75,;7.24,5.29,;8.58,6.06,;8.58,7.6,;9.91,5.29,;9.91,3.75,)| | ||
| Structure | 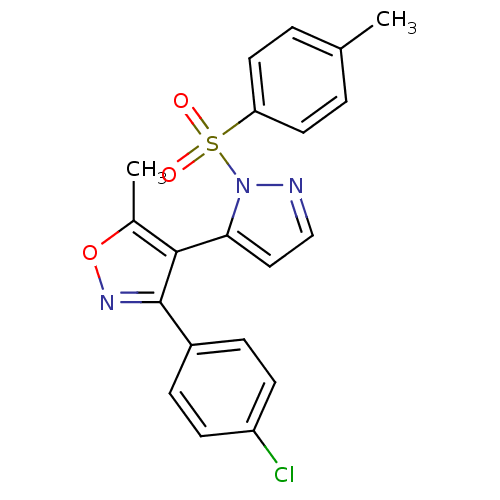
| ||