

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Vacuolar aminopeptidase 1 | ||
| Ligand | BDBM66062 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Dose Response of TOR pathway GFP-fusion proteins in Saccharomyes cerevisiae specifically LAP4 based on MLPCN hits | ||
| EC50 | 863±n/a nM | ||
| Citation |  PubChem, PC Dose Response of TOR pathway GFP-fusion proteins in Saccharomyes cerevisiae specifically LAP4 based on MLPCN hits PubChem Bioassay(2010)[AID] PubChem, PC Dose Response of TOR pathway GFP-fusion proteins in Saccharomyes cerevisiae specifically LAP4 based on MLPCN hits PubChem Bioassay(2010)[AID] | ||
| More Info.: | Get all data from this article, Solution Info, Assay Method | ||
| Vacuolar aminopeptidase 1 | |||
| Name: | Vacuolar aminopeptidase 1 | ||
| Synonyms: | AMPL_YEAST | APE1 | API | LAP4 | YSC1 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 57084.90 | ||
| Organism: | Saccharomyces cerevisiae | ||
| Description: | gi_486173 | ||
| Residue: | 514 | ||
| Sequence: |
| ||
| BDBM66062 | |||
| n/a | |||
| Name | BDBM66062 | ||
| Synonyms: | 2-keto-1H-pyridine-3-carboxylic acid [2-[1-(2-fluorophenyl)-2,5-dimethyl-pyrrol-3-yl]-2-keto-ethyl] ester | 2-oxo-1H-pyridine-3-carboxylic acid [2-[1-(2-fluorophenyl)-2,5-dimethyl-3-pyrrolyl]-2-oxoethyl] ester | MLS000761195 | SMR000365318 | [2-[1-(2-fluorophenyl)-2,5-dimethyl-pyrrol-3-yl]-2-oxidanylidene-ethyl] 2-oxidanylidene-1H-pyridine-3-carboxylate | [2-[1-(2-fluorophenyl)-2,5-dimethylpyrrol-3-yl]-2-oxoethyl] 2-oxo-1H-pyridine-3-carboxylate | cid_2472065 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H17FN2O4 | ||
| Mol. Mass. | 368.3584 | ||
| SMILES | Cc1cc(C(=O)COC(=O)c2ccc[nH]c2=O)c(C)n1-c1ccccc1F |(3.56,1.65,;5.09,1.81,;6.12,.66,;7.53,1.29,;8.87,.52,;10.2,1.29,;8.87,-1.02,;10.2,-1.79,;10.2,-3.33,;8.87,-4.1,;11.53,-4.1,;12.87,-3.33,;14.2,-4.1,;14.2,-5.64,;12.87,-6.41,;11.53,-5.64,;10.2,-6.41,;7.37,2.82,;8.52,3.85,;5.86,3.14,;5.24,4.55,;3.71,4.71,;3.08,6.12,;3.99,7.36,;5.52,7.2,;6.14,5.8,;7.67,5.63,)| | ||
| Structure | 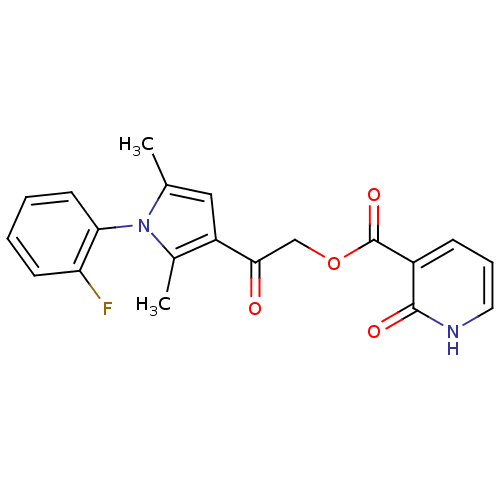
| ||