| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Protein RecA |
|---|
| Ligand | BDBM70736 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Intein inhibitors as potential Tuberculosis drugs |
|---|
| IC50 | 21750±n/a nM |
|---|
| Citation |  PubChem, PC Intein inhibitors as potential Tuberculosis drugs PubChem Bioassay(2010)[AID] PubChem, PC Intein inhibitors as potential Tuberculosis drugs PubChem Bioassay(2010)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Protein RecA |
|---|
| Name: | Protein RecA |
|---|
| Synonyms: | DNA recombination protein RecA | RECA_MYCTU | recA |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 85386.52 |
|---|
| Organism: | Mycobacterium tuberculosis H37Rv |
|---|
| Description: | gi_15609874 |
|---|
| Residue: | 790 |
|---|
| Sequence: | MTQTPDREKALELAVAQIEKSYGKGSVMRLGDEARQPISVIPTGSIALDVALGIGGLPRG
RVIEIYGPESSGKTTVALHAVANAQAAGGVAAFIDAEHALDPDYAKKLGVDTDSLLVSQP
DTGEQALEIADMLIRSGALDIVVIDSVAALVPRAELEGEMGDSHVGLQARLMSQALRKMT
GALNNSGTTAIFINQLRDKIGVMFGSPETTTGGKALKFYASVRMDVRRVETLKDGTNAVG
NRTRVKVVKNKCLAEGTRIFDPVTGTTHRIEDVVDGRKPIHVVAAAKDGTLHARPVVSWF
DQGTRDVIGLRIAGGAIVWATPDHKVLTEYGWRAAGELRKGDRVAQPRRFDGFGDSAPIP
ADHARLLGYLIGDGRDGWVGGKTPINFINVQRALIDDVTRIAATLGCAAHPQGRISLAIA
HRPGERNGVADLCQQAGIYGKLAWEKTIPNWFFEPDIAADIVGNLLFGLFESDGWVSREQ
TGALRVGYTTTSEQLAHQIHWLLLRFGVGSTVRDYDPTQKRPSIVNGRRIQSKRQVFEVR
ISGMDNVTAFAESVPMWGPRGAALIQAIPEATQGRRRGSQATYLAAEMTDAVLNYLDERG
VTAQEAAAMIGVASGDPRGGMKQVLGASRLRRDRVQALADALDDKFLHDMLAEELRYSVI
REVLPTRRARTFDLEVEELHTLVAEGVVVHNCSPPFKQAEFDILYGKGISREGSLIDMGV
DQGLIRKSGAWFTYEGEQLGQGKENARNFLVENADVADEIEKKIKEKLGIGAVVTDDPSN
DGVLPAPVDF
|
|
|
|---|
| BDBM70736 |
|---|
| n/a |
|---|
| Name | BDBM70736 |
|---|
| Synonyms: | 2-Chloro-N-(5-chloro-3-methyl-1-phenyl-1H-pyrazol-4-ylmethyl)-N-naphthalen-1-yl-acetamide | 2-chloranyl-N-[(5-chloranyl-3-methyl-1-phenyl-pyrazol-4-yl)methyl]-N-naphthalen-1-yl-ethanamide | 2-chloro-N-[(5-chloro-3-methyl-1-phenyl-4-pyrazolyl)methyl]-N-(1-naphthalenyl)acetamide | 2-chloro-N-[(5-chloro-3-methyl-1-phenyl-pyrazol-4-yl)methyl]-N-(1-naphthyl)acetamide | 2-chloro-N-[(5-chloro-3-methyl-1-phenylpyrazol-4-yl)methyl]-N-naphthalen-1-ylacetamide | MLS000566426 | SMR000177101 | cid_3134137 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H19Cl2N3O |
|---|
| Mol. Mass. | 424.322 |
|---|
| SMILES | Cc1nn(c(Cl)c1CN(C(=O)CCl)c1cccc2ccccc12)-c1ccccc1 |
|---|
| Structure | 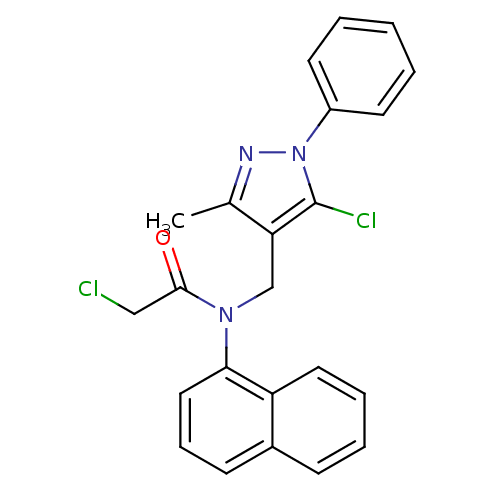
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Intein inhibitors as potential Tuberculosis drugs PubChem Bioassay(2010)[AID]
PubChem, PC Intein inhibitors as potential Tuberculosis drugs PubChem Bioassay(2010)[AID]