| Reaction Details |
|---|
 | Report a problem with these data |
| Target | D(1A) dopamine receptor |
|---|
| Ligand | BDBM50017696 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | 1780±n/a nM |
|---|
| Comments | PDSP_1533 |
|---|
| Citation |  Andersen, PH; Grønvald, FC; Jansen, JA A comparison between dopamine-stimulated adenylate cyclase and 3H-SCH 23390 binding in rat striatum. Life Sci37:1971-83 (1985) [PubMed] Article Andersen, PH; Grønvald, FC; Jansen, JA A comparison between dopamine-stimulated adenylate cyclase and 3H-SCH 23390 binding in rat striatum. Life Sci37:1971-83 (1985) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| D(1A) dopamine receptor |
|---|
| Name: | D(1A) dopamine receptor |
|---|
| Synonyms: | DOPAMINE D1 | DRD1_RAT | Dopamine D1 high | Dopamine D1 low | Dopamine receptor | Dopamine receptor D1 | Dopamine1-like | Drd1 | Drd1a |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 49429.75 |
|---|
| Organism: | RAT |
|---|
| Description: | P18901 |
|---|
| Residue: | 446 |
|---|
| Sequence: | MAPNTSTMDEAGLPAERDFSFRILTACFLSLLILSTLLGNTLVCAAVIRFRHLRSKVTNF
FVISLAVSDLLVAVLVMPWKAVAEIAGFWPLGPFCNIWVAFDIMCSTASILNLCVISVDR
YWAISSPFQYERKMTPKAAFILISVAWTLSVLISFIPVQLSWHKAKPTWPLDGNFTSLED
TEDDNCDTRLSRTYAISSSLISFYIPVAIMIVTYTSIYRIAQKQIRRISALERAAVHAKN
CQTTAGNGNPVECAQSESSFKMSFKRETKVLKTLSVIMGVFVCCWLPFFISNCMVPFCGS
EETQPFCIDSITFDVFVWFGWANSSLNPIIYAFNADFQKAFSTLLGCYRLCPTTNNAIET
VSINNNGAVVFSSHHEPRGSISKDCNLVYLIPHAVGSSEDLKKEEAGGIAKPLEKLSPAL
SVILDYDTDVSLEKIQPVTHSGQHST
|
|
|
|---|
| BDBM50017696 |
|---|
| n/a |
|---|
| Name | BDBM50017696 |
|---|
| Synonyms: | (2-dimethylamino-2-methyl)ethyl-N-dibenzoparathiazine | 10-(2-Dimethylaminopropyl)phenothiazine | 10-[2-(dimethylamino)propyl]phenothiazine | CHEMBL643 | N,N,alpha-trimethyl-10H-phenothiazine-10-ethanamine | N,N-dimethyl-1-(10H-phenothiazin-10-yl)propan-2-amine | PROMETHAZINE | med.21724, Compound 16 | proazamine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H20N2S |
|---|
| Mol. Mass. | 284.419 |
|---|
| SMILES | CC(CN1c2ccccc2Sc2ccccc12)N(C)C |
|---|
| Structure | 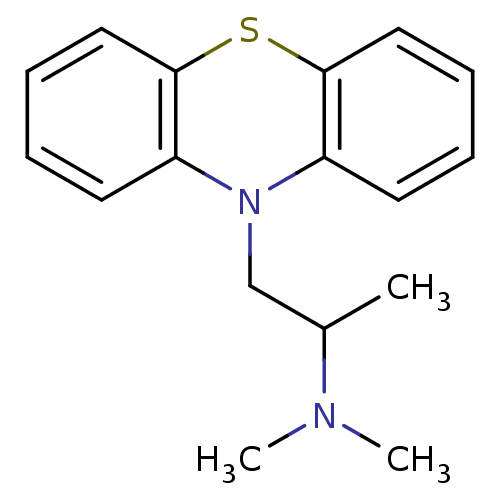
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Andersen, PH; Grønvald, FC; Jansen, JA A comparison between dopamine-stimulated adenylate cyclase and 3H-SCH 23390 binding in rat striatum. Life Sci37:1971-83 (1985) [PubMed] Article
Andersen, PH; Grønvald, FC; Jansen, JA A comparison between dopamine-stimulated adenylate cyclase and 3H-SCH 23390 binding in rat striatum. Life Sci37:1971-83 (1985) [PubMed] Article