| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histamine H1 receptor |
|---|
| Ligand | BDBM50005118 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | 8430±n/a nM |
|---|
| Comments | PDSP_1584 |
|---|
| Citation |  Hall, H; Sällemark, M; Jerning, E Effects of remoxipride and some related new substituted salicylamides on rat brain receptors. Acta Pharmacol Toxicol (Copenh)58:61-70 (1986) [PubMed] Article Hall, H; Sällemark, M; Jerning, E Effects of remoxipride and some related new substituted salicylamides on rat brain receptors. Acta Pharmacol Toxicol (Copenh)58:61-70 (1986) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Histamine H1 receptor |
|---|
| Name: | Histamine H1 receptor |
|---|
| Synonyms: | HISTAMINE H1 | HRH1_RAT | Hrh1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 55716.35 |
|---|
| Organism: | RAT |
|---|
| Description: | HISTAMINE H1 HRH1 RAT::P31390 |
|---|
| Residue: | 486 |
|---|
| Sequence: | MSFANTSSTFEDKMCEGNRTAMASPQLLPLVVVLSSISLVTVGLNLLVLYAVHSERKLHT
VGNLYIVSLSVADLIVGAVVMPMNILYLIMTKWSLGRPLCLFWLSMDYVASTASIFSVFI
LCIDRYRSVQQPLRYLRYRTKTRASATILGAWFFSFLWVIPILGWHHFMPPAPELREDKC
ETDFYNVTWFKIMTAIINFYLPTLLMLWFYVKIYKAVRRHCQHRQLTNGSLPSFSELKLR
SDDTKEGAKKPGRESPWGVLKRPSRDPSVGLDQKSTSEDPKMTSPTVFSQEGERETRPCF
RLDIMQKQSVAEGDVRGSKANDQALSQPKMDEQSLNTCRRISETSEDQTLVDQQSFSRTT
DSDTSIEPGPGRVKSRSGSNSGLDYIKITWKRLRSHSRQYVSGLHLNRERKAAKQLGFIM
AAFILCWIPYFIFFMVIAFCKSCCSEPMHMFTIWLGYINSTLNPLIYPLCNENFKKTFKK
ILHIRS
|
|
|
|---|
| BDBM50005118 |
|---|
| n/a |
|---|
| Name | BDBM50005118 |
|---|
| Synonyms: | (S)-3,5-Dichloro-N-(1-ethyl-pyrrolidin-2-ylmethyl)-2-hydroxy-6-methoxy-benzamide | (S)-3,5-dichloro-N-((1-ethylpyrrolidin-2-yl)methyl)-2-hydroxy-6-methoxybenzamide | (S)3,5-Dichloro-N-(1-ethyl-pyrrolidin-2-ylmethyl)-2-hydroxy-6-methoxy-benzamide | 3,5-Dichloro-N-((S)-1-ethyl-pyrrolidin-2-ylmethyl)-2-hydroxy-6-methoxy-benzamide | 3,5-Dichloro-N-(1-ethyl-pyrrolidin-2-ylmethyl)-2-hydroxy-6-methoxy-benzamide | 3,5-Dichloro-N-(1-ethyl-pyrrolidin-2-ylmethyl)-2-hydroxy-6-methoxy-benzamide (raclopride) | 3,5-Dichloro-N-(1-ethyl-pyrrolidin-2-ylmethyl)-2-hydroxy-6-methoxy-benzamide(Raclopride) | CHEMBL8809 | RACLOPRIDE | Raclopride;3,5-Dichloro-N-(1-ethyl-pyrrolidin-2-ylmethyl)-2-hydroxy-6-methoxy-benzamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H20Cl2N2O3 |
|---|
| Mol. Mass. | 347.237 |
|---|
| SMILES | CCN1CCC[C@H]1CNC(=O)c1c(O)c(Cl)cc(Cl)c1OC |
|---|
| Structure | 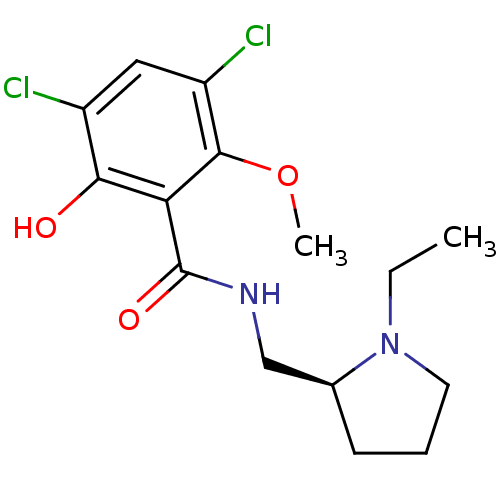
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hall, H; Sällemark, M; Jerning, E Effects of remoxipride and some related new substituted salicylamides on rat brain receptors. Acta Pharmacol Toxicol (Copenh)58:61-70 (1986) [PubMed] Article
Hall, H; Sällemark, M; Jerning, E Effects of remoxipride and some related new substituted salicylamides on rat brain receptors. Acta Pharmacol Toxicol (Copenh)58:61-70 (1986) [PubMed] Article