

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | 5-hydroxytryptamine receptor 3A | ||
| Ligand | BDBM50000492 | ||
| Substrate/Competitor | n/a | ||
| Ki | 0.32±n/a nM | ||
| Comments | PDSP_1991 | ||
| Citation |  Bonhaus, DW; Loury, DN; Jakeman, LB; To, Z; DeSouza, A; Eglen, RM; Wong, EH [3H]BIMU-1, a 5-hydroxytryptamine3 receptor ligand in NG-108 cells, selectively labels sigma-2 binding sites in guinea pig hippocampus. J Pharmacol Exp Ther267:961-70 (1993) [PubMed] Bonhaus, DW; Loury, DN; Jakeman, LB; To, Z; DeSouza, A; Eglen, RM; Wong, EH [3H]BIMU-1, a 5-hydroxytryptamine3 receptor ligand in NG-108 cells, selectively labels sigma-2 binding sites in guinea pig hippocampus. J Pharmacol Exp Ther267:961-70 (1993) [PubMed] | ||
| More Info.: | Get all data from this article | ||
| 5-hydroxytryptamine receptor 3A | |||
| Name: | 5-hydroxytryptamine receptor 3A | ||
| Synonyms: | 5-HT3 | 5-hydroxytryptamine receptor 3A | 5HT3A_RAT | 5ht3 | Htr3 | Htr3a | Serotonin (5-HT) receptor | Zacopride site-R | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 55428.70 | ||
| Organism: | RAT | ||
| Description: | 5-HT3 HTR3A RAT::P35563 | ||
| Residue: | 483 | ||
| Sequence: |
| ||
| BDBM50000492 | |||
| n/a | |||
| Name | BDBM50000492 | ||
| Synonyms: | (zacopride)4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide | 4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide | 4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide (Zacopride) | 4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide(zacopride) | CHEMBL18041 | HTR5A4-Amino-N-(1-aza-bicyclo[2.2.2]oct-3-yl)-5-chloro-2-methoxy-benzamide | ZACOPRIDE | ZACOPRIDE,R | ZACOPRIDE,RS | ZACOPRIDE,S | ||
| Type | Small organic molecule | ||
| Emp. Form. | C15H20ClN3O2 | ||
| Mol. Mass. | 309.791 | ||
| SMILES | COc1cc(N)c(Cl)cc1C(=O)NC1CN2CCC1CC2 |(27.19,-33.96,;28.52,-34.73,;28.53,-36.27,;27.2,-37.04,;27.2,-38.58,;25.86,-39.35,;28.53,-39.36,;28.53,-40.89,;29.87,-38.58,;29.86,-37.03,;31.19,-36.26,;31.19,-34.72,;32.53,-37.02,;33.86,-36.25,;35.2,-37.02,;36.52,-36.25,;36.52,-34.71,;35.19,-33.94,;33.85,-34.71,;34.61,-36.04,;35.74,-34.91,)| | ||
| Structure | 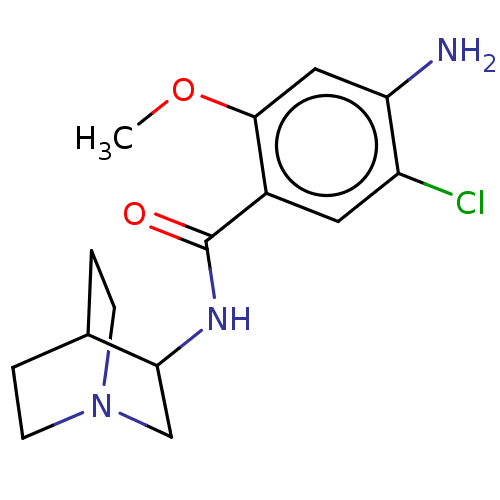
| ||