| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Apoptotic protease-activating factor 1 |
|---|
| Ligand | BDBM80290 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Dose Response confirmation of uHTS Activators of the Apaf-1 Pathway in Fluorescent format |
|---|
| EC50 | 4230±n/a nM |
|---|
| Citation |  PubChem, PC Dose Response confirmation of uHTS Activators of the Apaf-1 Pathway in Fluorescent format PubChem Bioassay(2011)[AID] PubChem, PC Dose Response confirmation of uHTS Activators of the Apaf-1 Pathway in Fluorescent format PubChem Bioassay(2011)[AID] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Apoptotic protease-activating factor 1 |
|---|
| Name: | Apoptotic protease-activating factor 1 |
|---|
| Synonyms: | APAF1 | APAF_HUMAN | Apoptotic peptidase activating factor 1 | KIAA0413 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 141834.40 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O14727 |
|---|
| Residue: | 1248 |
|---|
| Sequence: | MDAKARNCLLQHREALEKDIKTSYIMDHMISDGFLTISEEEKVRNEPTQQQRAAMLIKMI
LKKDNDSYVSFYNALLHEGYKDLAALLHDGIPVVSSSSGKDSVSGITSYVRTVLCEGGVP
QRPVVFVTRKKLVNAIQQKLSKLKGEPGWVTIHGMAGCGKSVLAAEAVRDHSLLEGCFPG
GVHWVSVGKQDKSGLLMKLQNLCTRLDQDESFSQRLPLNIEEAKDRLRILMLRKHPRSLL
ILDDVWDSWVLKAFDSQCQILLTTRDKSVTDSVMGPKYVVPVESSLGKEKGLEILSLFVN
MKKADLPEQAHSIIKECKGSPLVVSLIGALLRDFPNRWEYYLKQLQNKQFKRIRKSSSYD
YEALDEAMSISVEMLREDIKDYYTDLSILQKDVKVPTKVLCILWDMETEEVEDILQEFVN
KSLLFCDRNGKSFRYYLHDLQVDFLTEKNCSQLQDLHKKIITQFQRYHQPHTLSPDQEDC
MYWYNFLAYHMASAKMHKELCALMFSLDWIKAKTELVGPAHLIHEFVEYRHILDEKDCAV
SENFQEFLSLNGHLLGRQPFPNIVQLGLCEPETSEVYQQAKLQAKQEVDNGMLYLEWINK
KNITNLSRLVVRPHTDAVYHACFSEDGQRIASCGADKTLQVFKAETGEKLLEIKAHEDEV
LCCAFSTDDRFIATCSVDKKVKIWNSMTGELVHTYDEHSEQVNCCHFTNSSHHLLLATGS
SDCFLKLWDLNQKECRNTMFGHTNSVNHCRFSPDDKLLASCSADGTLKLWDATSANERKS
INVKQFFLNLEDPQEDMEVIVKCCSWSADGARIMVAAKNKIFLFDIHTSGLLGEIHTGHH
STIQYCDFSPQNHLAVVALSQYCVELWNTDSRSKVADCRGHLSWVHGVMFSPDGSSFLTS
SDDQTIRLWETKKVCKNSAVMLKQEVDVVFQENEVMVLAVDHIRRLQLINGRTGQIDYLT
EAQVSCCCLSPHLQYIAFGDENGAIEILELVNNRIFQSRFQHKKTVWHIQFTADEKTLIS
SSDDAEIQVWNWQLDKCIFLRGHQETVKDFRLLKNSRLLSWSFDGTVKVWNIITGNKEKD
FVCHQGTVLSCDISHDATKFSSTSADKTAKIWSFDLLLPLHELRGHNGCVRCSAFSVDST
LLATGDDNGEIRIWNVSNGELLHLCAPLSEEGAATHGGWVTDLCFSPDGKMLISAGGYIK
WWNVVTGESSQTFYTNGTNLKKIHVSPDFKTYVTVDNLGILYILQTLE
|
|
|
|---|
| BDBM80290 |
|---|
| n/a |
|---|
| Name | BDBM80290 |
|---|
| Synonyms: | (8R)-7-[2-(4-chloranylphenoxy)ethanoyl]-3-[3-(2-methylprop-2-enoylamino)phenyl]-1-oxa-2,7-diazaspiro[4.4]non-2-ene-8-carboxamide | (8R)-7-[2-(4-chlorophenoxy)-1-oxoethyl]-3-[3-[(2-methyl-1-oxoprop-2-enyl)amino]phenyl]-1-oxa-2,7-diazaspiro[4.4]non-2-ene-8-carboxamide | (8R)-7-[2-(4-chlorophenoxy)acetyl]-3-(3-methacrylamidophenyl)-1-oxa-2,7-diazaspiro[4.4]non-2-ene-8-carboxamide | (8R)-7-[2-(4-chlorophenoxy)acetyl]-3-[3-(2-methylprop-2-enoylamino)phenyl]-1-oxa-2,7-diazaspiro[4.4]non-2-ene-8-carboxamide | MLS000561957 | SMR000390644 | cid_16745478 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H25ClN4O5 |
|---|
| Mol. Mass. | 496.943 |
|---|
| SMILES | CC(=C)C(=O)Nc1cccc(c1)C1=NOC2(C[C@@H](N(C2)C(=O)COc2ccc(Cl)cc2)C(N)=O)C1 |t:13| |
|---|
| Structure | 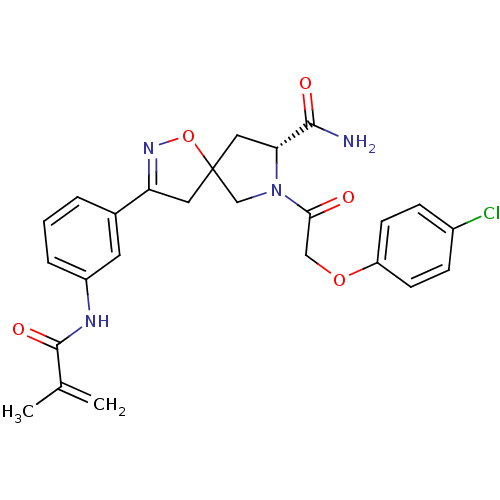
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Dose Response confirmation of uHTS Activators of the Apaf-1 Pathway in Fluorescent format PubChem Bioassay(2011)[AID]
PubChem, PC Dose Response confirmation of uHTS Activators of the Apaf-1 Pathway in Fluorescent format PubChem Bioassay(2011)[AID]