| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Aurora kinase C/Inner centromere protein |
|---|
| Ligand | BDBM50244953 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1674266 (CHEMBL4024295) |
|---|
| IC50 | 22±n/a nM |
|---|
| Citation |  Li, Y; Luo, X; Guo, Q; Nie, Y; Wang, T; Zhang, C; Huang, Z; Wang, X; Liu, Y; Chen, Y; Zheng, J; Yang, S; Fan, Y; Xiang, R Discovery of N1-(4-((7-Cyclopentyl-6-(dimethylcarbamoyl)-7 H-pyrrolo[2,3- d]pyrimidin-2-yl)amino)phenyl)- N8-hydroxyoctanediamide as a Novel Inhibitor Targeting Cyclin-dependent Kinase 4/9 (CDK4/9) and Histone Deacetlyase1 (HDAC1) against Malignant Cancer. J Med Chem61:3166-3192 (2018) [PubMed] Article Li, Y; Luo, X; Guo, Q; Nie, Y; Wang, T; Zhang, C; Huang, Z; Wang, X; Liu, Y; Chen, Y; Zheng, J; Yang, S; Fan, Y; Xiang, R Discovery of N1-(4-((7-Cyclopentyl-6-(dimethylcarbamoyl)-7 H-pyrrolo[2,3- d]pyrimidin-2-yl)amino)phenyl)- N8-hydroxyoctanediamide as a Novel Inhibitor Targeting Cyclin-dependent Kinase 4/9 (CDK4/9) and Histone Deacetlyase1 (HDAC1) against Malignant Cancer. J Med Chem61:3166-3192 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Aurora kinase C/Inner centromere protein |
|---|
| Name: | Aurora kinase C/Inner centromere protein |
|---|
| Synonyms: | Aurora-C/INCENP |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 1674266 |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | Aurora kinase C |
|---|
| Synonyms: | AIE2 | AIK3 | AIRK3 | ARK3 | AURKC | AURKC_HUMAN | Aurora Kinase C (Aurora-C) | Aurora kinase C | Aurora kinase C (AURKC) | Aurora-C | Aurora-C/INCENP | Aurora/Ipl1-related kinase 3 | Aurora/Ipl1/Eg2 protein 2 | STK13 | Serine/threonine-protein kinase 13 | Serine/threonine-protein kinase Aurora-C |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 35602.43 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Amino acid residues 1-309 were expressed as His-tagged fusion protein using baculovirus expression system. |
|---|
| Residue: | 309 |
|---|
| Sequence: | MSSPRAVVQLGKAQPAGEELATANQTAQQPSSPAMRRLTVDDFEIGRPLGKGKFGNVYLA
RLKESHFIVALKVLFKSQIEKEGLEHQLRREIEIQAHLQHPNILRLYNYFHDARRVYLIL
EYAPRGELYKELQKSEKLDEQRTATIIEELADALTYCHDKKVIHRDIKPENLLLGFRGEV
KIADFGWSVHTPSLRRKTMCGTLDYLPPEMIEGRTYDEKVDLWCIGVLCYELLVGYPPFE
SASHSETYRRILKVDVRFPLSMPLGARDLISRLLRYQPLERLPLAQILKHPWVQAHSRRV
LPPCAQMAS
|
|
|
|---|
| Component 2 |
| Name: | Inner centromere protein |
|---|
| Synonyms: | INCENP | INCE_HUMAN |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 105461.33 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9NQS7 |
|---|
| Residue: | 918 |
|---|
| Sequence: | MGTTAPGPIHLLELCDQKLMEFLCNMDNKDLVWLEEIQEEAERMFTREFSKEPELMPKTP
SQKNRRKKRRISYVQDENRDPIRRRLSRRKSRSSQLSSRRLRSKDSVEKLATVVGENGSV
LRRVTRAAAAAAAATMALAAPSSPTPESPTMLTKKPEDNHTQCQLVPVVEIGISERQNAE
QHVTQLMSTEPLPRTLSPTPASATAPTSQGIPTSDEESTPKKSKARILESITVSSLMATP
QDPKGQGVGTGRSASKLRIAQVSPGPRDSPAFPDSPWRERVLAPILPDNFSTPTGSRTDS
QSVRHSPIAPSSPSPQVLAQKYSLVAKQESVVRRASRRLAKKTAEEPAASGRIICHSYLE
RLLNVEVPQKVGSEQKEPPEEAEPVAAAEPEVPENNGNNSWPHNDTEIANSTPNPKPAAS
SPETPSAGQQEAKTDQADGPREPPQSARRKRSYKQAVSELDEEQHLEDEELQPPRSKTPS
SPCPASKVVRPLRTFLHTVQRNQMLMTPTSAPRSVMKSFIKRNTPLRMDPKCSFVEKERQ
RLENLRRKEEAEQLRRQKVEEDKRRRLEEVKLKREERLRKVLQARERVEQMKEEKKKQIE
QKFAQIDEKTEKAKEERLAEEKAKKKAAAKKMEEVEARRKQEEEARRLRWLQQEEEERRH
QELLQKKKEEEQERLRKAAEAKRLAEQREQERREQERREQERREQERREQERREQERQLA
EQERRREQERLQAERELQEREKALRLQKEQLQRELEEKKKKEEQQRLAERQLQEEQEKKA
KEAAGASKALNVTVDVQSPACTSYQMTPQGHRAPPKINPDNYGMDLNSDDSTDDEAHPRK
PIPTWARGTPLSQAIIHQYYHPPNLLELFGTILPLDLEDIFKKSKPRYHKRTSSAVWNSP
PLQGARVPSSLAYSLKKH
|
|
|
|---|
| BDBM50244953 |
|---|
| n/a |
|---|
| Name | BDBM50244953 |
|---|
| Synonyms: | CHEMBL4063500 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H37N7O4 |
|---|
| Mol. Mass. | 535.6379 |
|---|
| SMILES | CN(C)C(=O)c1cc2cnc(Nc3ccc(NC(=O)CCCCCCC(=O)NO)cc3)nc2n1C1CCCC1 |
|---|
| Structure | 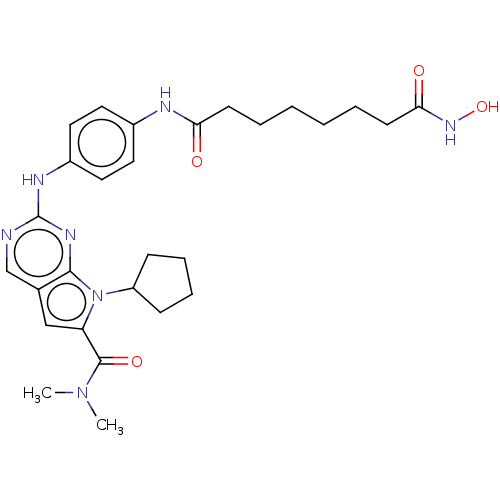
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Li, Y; Luo, X; Guo, Q; Nie, Y; Wang, T; Zhang, C; Huang, Z; Wang, X; Liu, Y; Chen, Y; Zheng, J; Yang, S; Fan, Y; Xiang, R Discovery of N1-(4-((7-Cyclopentyl-6-(dimethylcarbamoyl)-7 H-pyrrolo[2,3- d]pyrimidin-2-yl)amino)phenyl)- N8-hydroxyoctanediamide as a Novel Inhibitor Targeting Cyclin-dependent Kinase 4/9 (CDK4/9) and Histone Deacetlyase1 (HDAC1) against Malignant Cancer. J Med Chem61:3166-3192 (2018) [PubMed] Article
Li, Y; Luo, X; Guo, Q; Nie, Y; Wang, T; Zhang, C; Huang, Z; Wang, X; Liu, Y; Chen, Y; Zheng, J; Yang, S; Fan, Y; Xiang, R Discovery of N1-(4-((7-Cyclopentyl-6-(dimethylcarbamoyl)-7 H-pyrrolo[2,3- d]pyrimidin-2-yl)amino)phenyl)- N8-hydroxyoctanediamide as a Novel Inhibitor Targeting Cyclin-dependent Kinase 4/9 (CDK4/9) and Histone Deacetlyase1 (HDAC1) against Malignant Cancer. J Med Chem61:3166-3192 (2018) [PubMed] Article