| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prostaglandin G/H synthase 1/2 |
|---|
| Ligand | BDBM50000540 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_158283 (CHEMBL764933) |
|---|
| IC50 | 6000±n/a nM |
|---|
| Citation |  Malleron, JL; Roussel, G; Gueremy, G; Ponsinet, G; Robin, JL; Terlain, B; Tissieres, JM Penta- and hexadienoic acid derivatives: a novel series of 5-lipoxygenase inhibitors. J Med Chem33:2744-9 (1990) [PubMed] Malleron, JL; Roussel, G; Gueremy, G; Ponsinet, G; Robin, JL; Terlain, B; Tissieres, JM Penta- and hexadienoic acid derivatives: a novel series of 5-lipoxygenase inhibitors. J Med Chem33:2744-9 (1990) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prostaglandin G/H synthase 1/2 |
|---|
| Name: | Prostaglandin G/H synthase 1/2 |
|---|
| Synonyms: | Cyclooxygenase |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 1666355 |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | Prostaglandin G/H synthase 2 |
|---|
| Synonyms: | Cox-2 | Cox2 | Cyclooxygenase | PGH2_RAT | Ptgs2 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 69173.51 |
|---|
| Organism: | RAT |
|---|
| Description: | COX-2 0 RAT::P35355 |
|---|
| Residue: | 604 |
|---|
| Sequence: | MLFRAVLLCAALALSHAANPCCSNPCQNRGECMSIGFDQYKCDCTRTGFYGENCTTPEFL
TRIKLLLKPTPNTVHYILTHFKGVWNIVNNIPFLRNSIMRYVLTSRSHLIDSPPTYNVHY
GYKSWEAFSNLSYYTRALPPVADDCPTPMGVKGNKELPDSKEVLEKVLLRREFIPDPQGT
NMMFAFFAQHFTHQFFKTDQKRGPGFTRGLGHGVDLNHVYGETLDRQHKLRLFQDGKLKY
QVIGGEVYPPTVKDTQVDMIYPPHVPEHLRFAVGQEVFGLVPGLMMYATIWLREHNRVCD
ILKQEHPEWDDERLFQTSRLILIGETIKIVIEDYVQHLSGYHFKLKFDPELLFNQQFQYQ
NRIASEFNTLYHWHPLLPDTFNIEDQEYTFKQFLYNNSILLEHGLAHFVESFTRQIAGRV
AGGRNVPIAVQAVAKASIDQSREMKYQSLNEYRKRFSLKPYTSFEELTGEKEMAAELKAL
YHDIDAMELYPALLVEKPRPDAIFGETMVELGAPFSLKGLMGNPICSPQYWKPSTFGGEV
GFRIINTASIQSLICNNVKGCPFASFNVQDPQPTKTATINASASHSRLDDINPTVLIKRR
STEL
|
|
|
|---|
| Component 2 |
| Name: | Prostaglandin G/H synthase 1 |
|---|
| Synonyms: | Cox-1 | Cox1 | Cyclooxygenase | Cyclooxygenase-1 | PGH1_RAT | Prostaglandin G/H synthase (cyclooxygenase) | Ptgs1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 69033.56 |
|---|
| Organism: | RAT |
|---|
| Description: | COX-1 0 RAT::Q63921 |
|---|
| Residue: | 602 |
|---|
| Sequence: | MSRRSLSLQFPLLLLLLLLPPPPVLLTDAGVPSPVNPCCYYPCQNQGVCVRFGLDHYQCD
CTRTGYSGPNCTIPEIWTWLRSSLRPSPSFTHFLLTHGYWIWEFVNATFIREVLMRLVIT
VRSNLIPSPPTYNTAHDYISWESFSNVSYYTRILPSVPKDCPTPMGTKGKKQLPDIHLLA
QRLLLRREFIPGPQGTNVLFAFFAQHFTHQFFKTSGKMGPGFTKALGHGVDLGHIYGDSL
ERQYHLRLFKDGKLKYQVLDGEVYPPSVEQASVLMRYPPGVPPEKQMAVGQEVFGLLPGL
MLFSTIWLREHNRVCDLLKEEHPTWDDEQLFQTTRLILIGETIKIIIEEYVQHLSGYFLQ
LKFDPELLFRAQFQYRNRIALEFNHLYHWHPLMPDSFQVGSQEYSYEQFLFNTSMLVDYG
VEALVDAFSRQRAGRIGGGRNFDYHVLHVAEDVIKESREMRLQSFNEYRKRFGLKPYTSF
QEFTGEKEMAAELEELYGDIDALEFYPGLMLEKCQPNSLFGESMIEMGAPFSLKGLLGNP
ICSPEYWKPSTFGGDVGFNIVNTASLKKLVCLNTKTCPYVSFRVPDYPGDDGSVFVRPST
EL
|
|
|
|---|
| BDBM50000540 |
|---|
| n/a |
|---|
| Name | BDBM50000540 |
|---|
| Synonyms: | 1-(3-Trifluoromethyl-phenyl)-4,5-dihydro-1H-pyrazol-3-ylamine | 1-(3-Trifluoromethyl-phenyl)-4,5-dihydro-1H-pyrazol-3-ylamine (BW-755C) | 1-(3-Trifluoromethyl-phenyl)-4,5-dihydro-1H-pyrazol-3-ylamine(BW 755 C) | 1-(3-Trifluoromethyl-phenyl)-4,5-dihydro-1H-pyrazol-3-ylamine(BW 755C) | 1-(3-trifluoromethylphenyl)-4,5-dihydro-1H-3-pyrazolamine | BW-755C | CHEMBL274642 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H10F3N3 |
|---|
| Mol. Mass. | 229.2017 |
|---|
| SMILES | NC1=NN(CC1)c1cccc(c1)C(F)(F)F |t:1| |
|---|
| Structure | 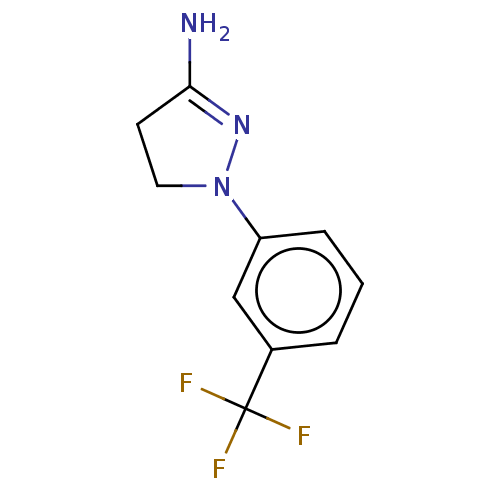
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Malleron, JL; Roussel, G; Gueremy, G; Ponsinet, G; Robin, JL; Terlain, B; Tissieres, JM Penta- and hexadienoic acid derivatives: a novel series of 5-lipoxygenase inhibitors. J Med Chem33:2744-9 (1990) [PubMed]
Malleron, JL; Roussel, G; Gueremy, G; Ponsinet, G; Robin, JL; Terlain, B; Tissieres, JM Penta- and hexadienoic acid derivatives: a novel series of 5-lipoxygenase inhibitors. J Med Chem33:2744-9 (1990) [PubMed]