| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-citrate synthase |
|---|
| Ligand | BDBM50018566 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_29530 (CHEMBL643517) |
|---|
| Ki | 56000000±n/a nM |
|---|
| Citation |  Bar-Tana, J; Ben-Shoshan, S; Blum, J; Migron, Y; Hertz, R; Pill, J; Rose-Khan, G; Witte, EC Synthesis and hypolipidemic and antidiabetogenic activities of beta,beta,beta',beta'-tetrasubstituted, long-chain dioic acids. J Med Chem32:2072-84 (1989) [PubMed] Bar-Tana, J; Ben-Shoshan, S; Blum, J; Migron, Y; Hertz, R; Pill, J; Rose-Khan, G; Witte, EC Synthesis and hypolipidemic and antidiabetogenic activities of beta,beta,beta',beta'-tetrasubstituted, long-chain dioic acids. J Med Chem32:2072-84 (1989) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| ATP-citrate synthase |
|---|
| Name: | ATP-citrate synthase |
|---|
| Synonyms: | ACLY_RAT | Acly |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 120645.65 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_857791 |
|---|
| Residue: | 1100 |
|---|
| Sequence: | MSAKAISEQTGKELLYKYICTTSAIQNRFKYARVTPDTDWAHLLQDHPWLLSQSLVVKPD
QLIKRRGKLGLVGVNLSLDGVKSWLKPRLGHEATVGKAKGFLKNFLIEPFVPHSQAEEFY
VCIYATREGDYVLFHHEGGVDVGDVDTKAQKLLVGVDEKLNAEDIKRHLLVHAPEDKKEI
LASFISGLFNFYEDLYFTYLEINPLVVTKDGVYILDLAAKVDATADYICKVKWGDIEFPP
PFGREAYPEEAYIADLDAKSGASLKLTLLNPKGRIWTMVAGGGASVVYSDTICDLGGVNE
LANYGEYSGAPSEQQTYDYAKTILSLMTREKHPDGKILIIGGSIANFTNVAATFKGIVRA
IRDYQGSLKEHEVTIFVRRGGPNYQEGLRVMGEVGKTTGIPIHVFGTETHMTAIVGMAWA
PAIPNQPPTAAHTANFLLNASGSTSTPAPSRTASFSESRADEVAPAKKAKPAMPQDSVPS
PRSLQGKSATLFSRHTKAIVWGMQTRAVQGMLDFDYVCSRDEPSVAAMVYPFTGDHKQKF
YWGHKEILIPVFKNMADAMKKHPEVDVLINFASLRSAYDSTMETMNYAQIRTIAIIAEGI
PEALTRKLIKKADQKGVTIIGPATVGGIKPGCFKIGNTGGMLDNILASKLYRPGSVAYVS
RSGGMSNELNNIISRTTDGVYEGVAIGGDRYPGSTFMDHVLRYQDTPGVKMIVVLGEIGG
TEEYKICRGIKEGRLTKPVVCWCIGTCATMFSSEVQFGHAGACANQASETAVAKNQALKE
AGVFVPRSFDELGEIIQSVYEDLVAKGAIVPAQEVPPPTVPMDYSWARELGLIRKPASFM
TSICDERGQELIYAGMPITEVFKEEMGIGGVLGLLWFQRRLPKYSCQFIEMCLMVTADHG
PAVSGAHNTIICARAGKDLVSSLTSGLLTIGDRFGGALDAAAKMFSKAFDSGIIPMEFVN
KMKKEGKLIMGIGHRVKSINNPDMRVQILKDFVKQHFPATPLLDYALEVEKITTSKKPNL
ILNVDGFIGVAFVDMLRNCGSFTREEADEYVDIGALNGVFVLGRSMGFIGHYLDQKRLKQ
GLYRHPWDDISYVLPEHMSM
|
|
|
|---|
| BDBM50018566 |
|---|
| n/a |
|---|
| Name | BDBM50018566 |
|---|
| Synonyms: | 3,3,12,12-Tetramethyl-tetradecanedioic acid | CHEMBL64960 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H34O4 |
|---|
| Mol. Mass. | 314.4602 |
|---|
| SMILES | CC(C)(CCCCCCCCC(C)(C)CC(O)=O)CC(O)=O |
|---|
| Structure | 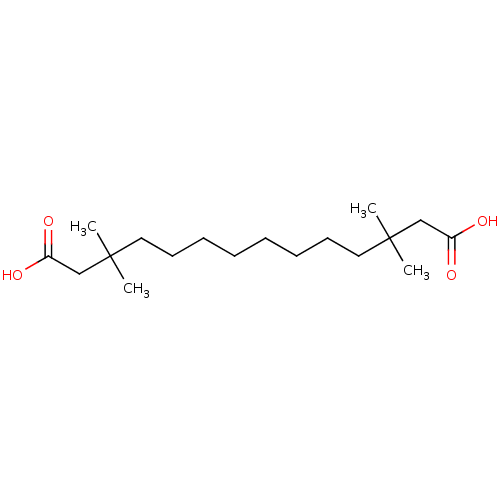
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bar-Tana, J; Ben-Shoshan, S; Blum, J; Migron, Y; Hertz, R; Pill, J; Rose-Khan, G; Witte, EC Synthesis and hypolipidemic and antidiabetogenic activities of beta,beta,beta',beta'-tetrasubstituted, long-chain dioic acids. J Med Chem32:2072-84 (1989) [PubMed]
Bar-Tana, J; Ben-Shoshan, S; Blum, J; Migron, Y; Hertz, R; Pill, J; Rose-Khan, G; Witte, EC Synthesis and hypolipidemic and antidiabetogenic activities of beta,beta,beta',beta'-tetrasubstituted, long-chain dioic acids. J Med Chem32:2072-84 (1989) [PubMed]