

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Tyrosine-protein phosphatase non-receptor type 1 | ||
| Ligand | BDBM50362839 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1754336 (CHEMBL4189096) | ||
| IC50 | 37900±n/a nM | ||
| Citation |  Zhang, J; Sasaki, T; Li, W; Nagata, K; Higai, K; Feng, F; Wang, J; Cheng, M; Koike, K Identification of caffeoylquinic acid derivatives as natural protein tyrosine phosphatase 1B inhibitors from Artemisia princeps. Bioorg Med Chem Lett28:1194-1197 (2018) [PubMed] Article Zhang, J; Sasaki, T; Li, W; Nagata, K; Higai, K; Feng, F; Wang, J; Cheng, M; Koike, K Identification of caffeoylquinic acid derivatives as natural protein tyrosine phosphatase 1B inhibitors from Artemisia princeps. Bioorg Med Chem Lett28:1194-1197 (2018) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Tyrosine-protein phosphatase non-receptor type 1 | |||
| Name: | Tyrosine-protein phosphatase non-receptor type 1 | ||
| Synonyms: | PTN1_HUMAN | PTP1B | PTPN1 | Protein tyrosine phosphatase 1B (PTP1B) | Protein tyrosine phosphatase-1B (PTP1B) | Protein-tyrosine phosphatase 1B | Protein-tyrosine phosphatase 1B (PTP1B) | Tyrosine-protein phosphatase non-receptor type 1 | Tyrosine-protein phosphatase non-receptor type 1 (PTP1B) | ||
| Type: | Protein phosphatase | ||
| Mol. Mass.: | 49963.76 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Human recombinant GST-fusion PTP1B (1-435). | ||
| Residue: | 435 | ||
| Sequence: |
| ||
| BDBM50362839 | |||
| n/a | |||
| Name | BDBM50362839 | ||
| Synonyms: | CHEMBL249447 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H24O12 | ||
| Mol. Mass. | 516.4509 | ||
| SMILES | O[C@H]1[C@@H](C[C@@](O)(C[C@H]1OC(=O)\C=C\c1ccc(O)c(O)c1)C(O)=O)OC(=O)\C=C\c1ccc(O)c(O)c1 |r,wU:2.25,4.22,wD:1.0,7.8,4.4,(-1.06,-23.53,;-1.06,-21.99,;.29,-21.22,;.29,-19.66,;-1.06,-18.89,;.22,-18.03,;-2.39,-19.66,;-2.39,-21.22,;-3.73,-21.99,;-3.73,-23.53,;-2.39,-24.3,;-5.06,-24.3,;-5.06,-25.84,;-6.39,-26.61,;-7.73,-25.84,;-9.06,-26.61,;-9.06,-28.15,;-10.39,-28.92,;-7.72,-28.92,;-7.71,-30.46,;-6.39,-28.15,;-1.91,-17.6,;-.95,-16.4,;-3.45,-17.53,;1.62,-21.99,;1.61,-23.53,;.28,-24.3,;2.95,-24.3,;2.94,-25.84,;4.27,-26.62,;4.26,-28.16,;5.59,-28.93,;6.93,-28.16,;8.26,-28.93,;6.93,-26.61,;8.26,-25.84,;5.6,-25.85,)| | ||
| Structure | 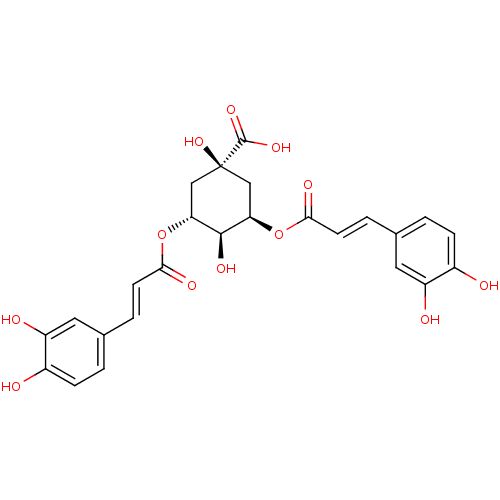
| ||