| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-secretase 1 |
|---|
| Ligand | BDBM50398264 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1759779 (CHEMBL4194787) |
|---|
| Ki | 26±n/a nM |
|---|
| Citation |  Young, RJ; Leeson, PD Mapping the Efficiency and Physicochemical Trajectories of Successful Optimizations. J Med Chem61:6421-6467 (2018) [PubMed] Article Young, RJ; Leeson, PD Mapping the Efficiency and Physicochemical Trajectories of Successful Optimizations. J Med Chem61:6421-6467 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Beta-secretase 1 |
|---|
| Name: | Beta-secretase 1 |
|---|
| Synonyms: | ASP2 | Asp 2 | Aspartyl protease 2 | BACE | BACE1 | BACE1_HUMAN | Beta-secretase (BACE) | Beta-secretase 1 | Beta-secretase 1 (BACE 1) | Beta-secretase 1 (BACE-1) | Beta-secretase 1 (BACE1) | Beta-site APP cleaving enzyme 1 | Beta-site amyloid precursor protein cleaving enzyme 1 | KIAA1149 | Memapsin-2 | Membrane-associated aspartic protease 2 | beta-Secretase (BACE-1) | beta-Secretase (BACE1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 55755.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P56817 |
|---|
| Residue: | 501 |
|---|
| Sequence: | MAQALPWLLLWMGAGVLPAHGTQHGIRLPLRSGLGGAPLGLRLPRETDEEPEEPGRRGSF
VEMVDNLRGKSGQGYYVEMTVGSPPQTLNILVDTGSSNFAVGAAPHPFLHRYYQRQLSST
YRDLRKGVYVPYTQGKWEGELGTDLVSIPHGPNVTVRANIAAITESDKFFINGSNWEGIL
GLAYAEIARPDDSLEPFFDSLVKQTHVPNLFSLQLCGAGFPLNQSEVLASVGGSMIIGGI
DHSLYTGSLWYTPIRREWYYEVIIVRVEINGQDLKMDCKEYNYDKSIVDSGTTNLRLPKK
VFEAAVKSIKAASSTEKFPDGFWLGEQLVCWQAGTTPWNIFPVISLYLMGEVTNQSFRIT
ILPQQYLRPVEDVATSQDDCYKFAISQSSTGTVMGAVIMEGFYVVFDRARKRIGFAVSAC
HVHDEFRTAAVEGPFVTLDMEDCGYNIPQTDESTLMTIAYVMAAICALFMLPLCLMVCQW
RCLRCLRQQHDDFADDISLLK
|
|
|
|---|
| BDBM50398264 |
|---|
| n/a |
|---|
| Name | BDBM50398264 |
|---|
| Synonyms: | CHEMBL2177913 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H16F3N5 |
|---|
| Mol. Mass. | 431.4125 |
|---|
| SMILES | NC1=N[C@@](c2cccc(F)c12)(c1cccc(c1)-c1cncnc1)c1ccnc(c1)C(F)F |r,t:1| |
|---|
| Structure | 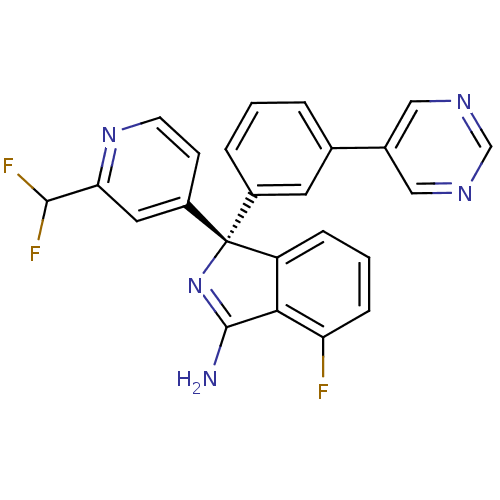
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Young, RJ; Leeson, PD Mapping the Efficiency and Physicochemical Trajectories of Successful Optimizations. J Med Chem61:6421-6467 (2018) [PubMed] Article
Young, RJ; Leeson, PD Mapping the Efficiency and Physicochemical Trajectories of Successful Optimizations. J Med Chem61:6421-6467 (2018) [PubMed] Article