

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | E3 ubiquitin-protein ligase XIAP | ||
| Ligand | BDBM50465643 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1785296 (CHEMBL4256813) | ||
| IC50 | 5944±n/a nM | ||
| Citation |  Baggio, C; Gambini, L; Udompholkul, P; Salem, AF; Aronson, A; Dona, A; Troadec, E; Pichiorri, F; Pellecchia, M Design of Potent pan-IAP and Lys-Covalent XIAP Selective Inhibitors Using a Thermodynamics Driven Approach. J Med Chem61:6350-6363 (2018) [PubMed] Article Baggio, C; Gambini, L; Udompholkul, P; Salem, AF; Aronson, A; Dona, A; Troadec, E; Pichiorri, F; Pellecchia, M Design of Potent pan-IAP and Lys-Covalent XIAP Selective Inhibitors Using a Thermodynamics Driven Approach. J Med Chem61:6350-6363 (2018) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| E3 ubiquitin-protein ligase XIAP | |||
| Name: | E3 ubiquitin-protein ligase XIAP | ||
| Synonyms: | API3 | BIRC4 | E3 ubiquitin-protein ligase XIAP | IAP3 | Inhibitor of apoptosis protein 3 | Inhibitor of apoptosis protein 3 (XIAP) | X-linked inhibitor of apoptosis | X-linked inhibitor of apoptosis protein (XIAP) | XIAP | XIAP_HUMAN | ||
| Type: | Protein | ||
| Mol. Mass.: | 56685.27 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P98170 | ||
| Residue: | 497 | ||
| Sequence: |
| ||
| BDBM50465643 | |||
| n/a | |||
| Name | BDBM50465643 | ||
| Synonyms: | CHEMBL4284657 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H32N8O2 | ||
| Mol. Mass. | 476.574 | ||
| SMILES | CN[C@@H](C)C(=O)N[C@@H](Cc1ccc(cc1)N=C(N)N)C(=O)N1CCCC1c1nc2ccccc2[nH]1 |r,wU:7.7,wD:2.2,(46.71,-8.64,;48.04,-7.87,;49.37,-8.64,;49.37,-10.18,;50.71,-7.87,;50.71,-6.33,;52.04,-8.64,;53.37,-7.87,;53.37,-6.33,;52.27,-5.23,;52.69,-3.72,;51.59,-2.62,;50.08,-3.02,;49.68,-4.53,;50.78,-5.63,;48.99,-1.93,;47.5,-2.33,;46.42,-1.24,;47.1,-3.81,;54.71,-8.64,;54.71,-10.18,;56.05,-7.87,;56.74,-6.5,;58.27,-6.74,;58.51,-8.26,;57.13,-8.96,;56.9,-10.48,;55.53,-11.18,;55.77,-12.69,;54.8,-13.88,;55.35,-15.32,;56.87,-15.56,;57.84,-14.37,;57.29,-12.94,;57.99,-11.57,)| | ||
| Structure | 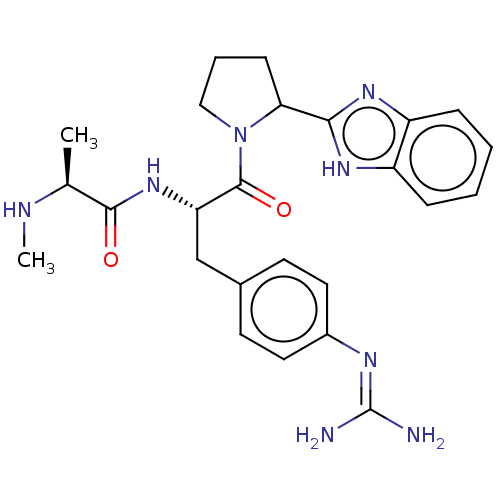
| ||