

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mitogen-activated protein kinase 10 | ||
| Ligand | BDBM50364378 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1800095 (CHEMBL4272387) | ||
| IC50 | 6.0±n/a nM | ||
| Citation |  Juillerat-Jeanneret, L; Aubert, JD; Mikulic, J; Golshayan, D Fibrogenic Disorders in Human Diseases: From Inflammation to Organ Dysfunction. J Med Chem61:9811-9840 (2018) [PubMed] Article Juillerat-Jeanneret, L; Aubert, JD; Mikulic, J; Golshayan, D Fibrogenic Disorders in Human Diseases: From Inflammation to Organ Dysfunction. J Med Chem61:9811-9840 (2018) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mitogen-activated protein kinase 10 | |||
| Name: | Mitogen-activated protein kinase 10 | ||
| Synonyms: | JNK3 | JNK3A | MAP kinase p49 3F12 | MAPK10 | MK10_HUMAN | Mitogen-Activated Protein Kinase 10 (JNK3) | Mitogen-activated protein kinase 10 (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12) | Mitogen-activated protein kinase 10/Receptor-interacting serine/threonine-protein kinase 1 | PRKM10 | SAPK1B | Stress-activated protein kinase JNK3 | c-Jun N-terminal kinase 3 (JNK3) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 52586.89 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 464 | ||
| Sequence: |
| ||
| BDBM50364378 | |||
| n/a | |||
| Name | BDBM50364378 | ||
| Synonyms: | CHEMBL1950289 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C21H23F3N6O2 | ||
| Mol. Mass. | 448.4415 | ||
| SMILES | O[C@H]1CC[C@@H](CC1)Nc1ncc2nc(Nc3c(F)cc(F)cc3F)n([C@H]3CCOC3)c2n1 |r,wU:1.0,wD:4.7,25.26,(-6.26,-6.49,;-5.49,-5.16,;-6.25,-3.82,;-5.49,-2.5,;-3.95,-2.49,;-3.17,-3.82,;-3.94,-5.16,;-3.19,-1.16,;-1.65,-1.15,;-.89,.17,;.66,.18,;1.42,-1.15,;2.92,-1.46,;3.09,-2.98,;4.42,-3.75,;5.75,-2.97,;5.74,-1.45,;4.4,-.69,;7.06,-.67,;8.41,-1.43,;9.74,-.66,;8.41,-2.97,;7.08,-3.75,;7.08,-5.29,;1.69,-3.61,;1.23,-5.08,;-.23,-5.56,;-.23,-7.1,;1.24,-7.57,;2.14,-6.32,;.66,-2.48,;-.88,-2.48,)| | ||
| Structure | 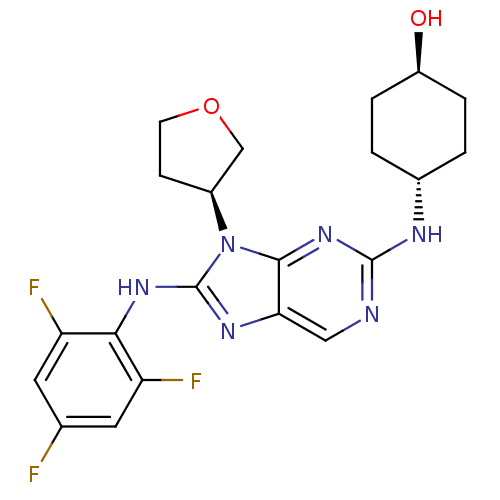
| ||