| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Protease |
|---|
| Ligand | BDBM580 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_469686 (CHEMBL946814) |
|---|
| Ki | 0.035±n/a nM |
|---|
| Citation |  Mimoto, T; Nojima, S; Terashima, K; Takaku, H; Shintani, M; Hayashi, H Structure-activity relationships of novel HIV-1 protease inhibitors containing the 3-amino-2-chlorobenzoyl-allophenylnorstatine structure. Bioorg Med Chem16:1299-308 (2008) [PubMed] Article Mimoto, T; Nojima, S; Terashima, K; Takaku, H; Shintani, M; Hayashi, H Structure-activity relationships of novel HIV-1 protease inhibitors containing the 3-amino-2-chlorobenzoyl-allophenylnorstatine structure. Bioorg Med Chem16:1299-308 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Protease |
|---|
| Name: | Protease |
|---|
| Synonyms: | n/a |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 10904.79 |
|---|
| Organism: | Human immunodeficiency virus 1 (HIV-1) |
|---|
| Description: | Q9YQ12 |
|---|
| Residue: | 99 |
|---|
| Sequence: | PQITLWQRPFVTIKIEGQLKEALLDTGADDTVLEEMNLPGRWKPKMIGGIGGFIKVRQYD
QIVIEICGKKAIGTVLVGPTPVNIIGRNLLTQIGCTLNF
|
|
|
|---|
| BDBM580 |
|---|
| n/a |
|---|
| Name | BDBM580 |
|---|
| Synonyms: | (4R)-3-[(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylphenyl)formamido]-4-phenylbutanoyl]-5,5-dimethyl-N-[(2-methylphenyl)methyl]-1,3-thiazolidine-4-carboxamide | AG1776 | BDBM50288366 | JE-2147 | KNI-764 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C32H37N3O5S |
|---|
| Mol. Mass. | 575.718 |
|---|
| SMILES | Cc1ccccc1CNC(=O)[C@H]1N(CSC1(C)C)C(=O)[C@@H](O)[C@H](Cc1ccccc1)NC(=O)c1cccc(O)c1C |r| |
|---|
| Structure | 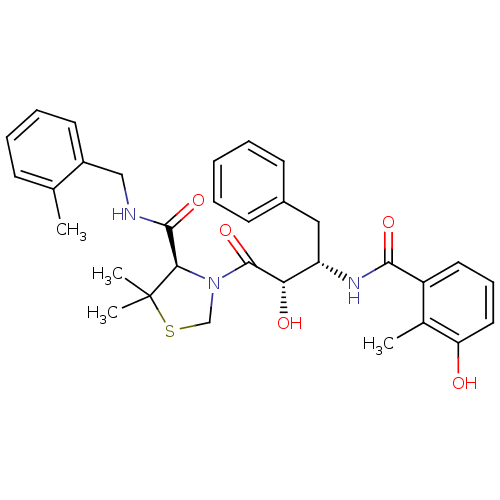
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Mimoto, T; Nojima, S; Terashima, K; Takaku, H; Shintani, M; Hayashi, H Structure-activity relationships of novel HIV-1 protease inhibitors containing the 3-amino-2-chlorobenzoyl-allophenylnorstatine structure. Bioorg Med Chem16:1299-308 (2008) [PubMed] Article
Mimoto, T; Nojima, S; Terashima, K; Takaku, H; Shintani, M; Hayashi, H Structure-activity relationships of novel HIV-1 protease inhibitors containing the 3-amino-2-chlorobenzoyl-allophenylnorstatine structure. Bioorg Med Chem16:1299-308 (2008) [PubMed] Article