| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Ligand | BDBM50200120 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_478364 (CHEMBL931316) |
|---|
| Ki | 5.0±n/a nM |
|---|
| Citation |  Barb, AW; Jiang, L; Raetz, CR; Zhou, P Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: Time-dependent inhibition and specificity in ligand binding. Proc Natl Acad Sci U S A104:18433-8 (2007) [PubMed] Article Barb, AW; Jiang, L; Raetz, CR; Zhou, P Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: Time-dependent inhibition and specificity in ligand binding. Proc Natl Acad Sci U S A104:18433-8 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Name: | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Synonyms: | LPXC_ECOLI | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LpxC) | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LxpC) | UDP-3-O-acyl-GlcNAc deacetylase | asmB | envA | lpxC |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 33952.00 |
|---|
| Organism: | Escherichia coli |
|---|
| Description: | P0A725 |
|---|
| Residue: | 305 |
|---|
| Sequence: | MIKQRTLKRIVQATGVGLHTGKKVTLTLRPAPANTGVIYRRTDLNPPVDFPADAKSVRDT
MLCTCLVNEHDVRISTVEHLNAALAGLGIDNIVIEVNAPEIPIMDGSAAPFVYLLLDAGI
DELNCAKKFVRIKETVRVEDGDKWAEFKPYNGFSLDFTIDFNHPAIDSSNQRYAMNFSAD
AFMRQISRARTFGFMRDIEYLQSRGLCLGGSFDCAIVVDDYRVLNEDGLRFEDEFVRHKM
LDAIGDLFMCGHNIIGAFTAYKSGHALNNKLLQAVLAKQEAWEYVTFQDDAELPLAFKAP
SAVLA
|
|
|
|---|
| BDBM50200120 |
|---|
| n/a |
|---|
| Name | BDBM50200120 |
|---|
| Synonyms: | CHEMBL260091 | CHIR-090 | US10875832, Compound ChIR-090 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H27N3O5 |
|---|
| Mol. Mass. | 437.4883 |
|---|
| SMILES | C[C@@H](O)[C@H](NC(=O)c1ccc(cc1)C#Cc1ccc(CN2CCOCC2)cc1)C(=O)NO |r| |
|---|
| Structure | 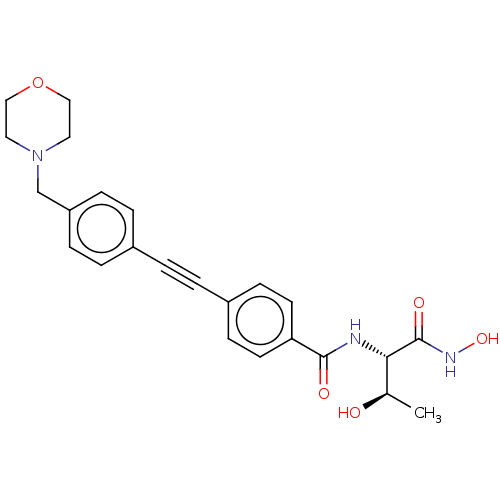
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Barb, AW; Jiang, L; Raetz, CR; Zhou, P Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: Time-dependent inhibition and specificity in ligand binding. Proc Natl Acad Sci U S A104:18433-8 (2007) [PubMed] Article
Barb, AW; Jiang, L; Raetz, CR; Zhou, P Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: Time-dependent inhibition and specificity in ligand binding. Proc Natl Acad Sci U S A104:18433-8 (2007) [PubMed] Article