| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cystic fibrosis transmembrane conductance regulator |
|---|
| Ligand | BDBM100205 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_632278 (CHEMBL1110140) |
|---|
| EC50 | 700±n/a nM |
|---|
| Citation |  Ye, L; Knapp, JM; Sangwung, P; Fettinger, JC; Verkman, AS; Kurth, MJ Pyrazolylthiazole as DeltaF508-cystic fibrosis transmembrane conductance regulator correctors with improved hydrophilicity compared to bithiazoles. J Med Chem53:3772-81 (2010) [PubMed] Article Ye, L; Knapp, JM; Sangwung, P; Fettinger, JC; Verkman, AS; Kurth, MJ Pyrazolylthiazole as DeltaF508-cystic fibrosis transmembrane conductance regulator correctors with improved hydrophilicity compared to bithiazoles. J Med Chem53:3772-81 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cystic fibrosis transmembrane conductance regulator |
|---|
| Name: | Cystic fibrosis transmembrane conductance regulator |
|---|
| Synonyms: | ABCC7 | ATP-binding cassette sub-family C member 7 | CFTR | CFTR_HUMAN | Channel conductance-controlling ATPase | cAMP-dependent chloride channel |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 168171.30 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | gi_90421313 |
|---|
| Residue: | 1480 |
|---|
| Sequence: | MQRSPLEKASVVSKLFFSWTRPILRKGYRQRLELSDIYQIPSVDSADNLSEKLEREWDRE
LASKKNPKLINALRRCFFWRFMFYGIFLYLGEVTKAVQPLLLGRIIASYDPDNKEERSIA
IYLGIGLCLLFIVRTLLLHPAIFGLHHIGMQMRIAMFSLIYKKTLKLSSRVLDKISIGQL
VSLLSNNLNKFDEGLALAHFVWIAPLQVALLMGLIWELLQASAFCGLGFLIVLALFQAGL
GRMMMKYRDQRAGKISERLVITSEMIENIQSVKAYCWEEAMEKMIENLRQTELKLTRKAA
YVRYFNSSAFFFSGFFVVFLSVLPYALIKGIILRKIFTTISFCIVLRMAVTRQFPWAVQT
WYDSLGAINKIQDFLQKQEYKTLEYNLTTTEVVMENVTAFWEEGFGELFEKAKQNNNNRK
TSNGDDSLFFSNFSLLGTPVLKDINFKIERGQLLAVAGSTGAGKTSLLMVIMGELEPSEG
KIKHSGRISFCSQFSWIMPGTIKENIIFGVSYDEYRYRSVIKACQLEEDISKFAEKDNIV
LGEGGITLSGGQRARISLARAVYKDADLYLLDSPFGYLDVLTEKEIFESCVCKLMANKTR
ILVTSKMEHLKKADKILILHEGSSYFYGTFSELQNLQPDFSSKLMGCDSFDQFSAERRNS
ILTETLHRFSLEGDAPVSWTETKKQSFKQTGEFGEKRKNSILNPINSIRKFSIVQKTPLQ
MNGIEEDSDEPLERRLSLVPDSEQGEAILPRISVISTGPTLQARRRQSVLNLMTHSVNQG
QNIHRKTTASTRKVSLAPQANLTELDIYSRRLSQETGLEISEEINEEDLKECFFDDMESI
PAVTTWNTYLRYITVHKSLIFVLIWCLVIFLAEVAASLVVLWLLGNTPLQDKGNSTHSRN
NSYAVIITSTSSYYVFYIYVGVADTLLAMGFFRGLPLVHTLITVSKILHHKMLHSVLQAP
MSTLNTLKAGGILNRFSKDIAILDDLLPLTIFDFIQLLLIVIGAIAVVAVLQPYIFVATV
PVIVAFIMLRAYFLQTSQQLKQLESEGRSPIFTHLVTSLKGLWTLRAFGRQPYFETLFHK
ALNLHTANWFLYLSTLRWFQMRIEMIFVIFFIAVTFISILTTGEGEGRVGIILTLAMNIM
STLQWAVNSSIDVDSLMRSVSRVFKFIDMPTEGKPTKSTKPYKNGQLSKVMIIENSHVKK
DDIWPSGGQMTVKDLTAKYTEGGNAILENISFSISPGQRVGLLGRTGSGKSTLLSAFLRL
LNTEGEIQIDGVSWDSITLQQWRKAFGVIPQKVFIFSGTFRKNLDPYEQWSDQEIWKVAD
EVGLRSVIEQFPGKLDFVLVDGGCVLSHGHKQLMCLARSVLSKAKILLLDEPSAHLDPVT
YQIIRRTLKQAFADCTVILCEHRIEAMLECQQFLVIEENKVRQYDSIQKLLNERSLFRQA
ISPSDRVKLFPHRNSSKCKSKPQIAALKEETEEEVQDTRL
|
|
|
|---|
| BDBM100205 |
|---|
| n/a |
|---|
| Name | BDBM100205 |
|---|
| Synonyms: | MLS000561583 | N-[5-[2-(5-chloro-2-methoxy-anilino)thiazol-4-yl]-4-methyl-thiazol-2-yl]-2,2-dimethyl-propionamide | N-[5-[2-(5-chloro-2-methoxyanilino)-1,3-thiazol-4-yl]-4-methyl-1,3-thiazol-2-yl]-2,2-dimethylpropanamide | N-[5-[2-(5-chloro-2-methoxyanilino)-4-thiazolyl]-4-methyl-2-thiazolyl]-2,2-dimethylpropanamide | N-[5-[2-[(5-chloranyl-2-methoxy-phenyl)amino]-1,3-thiazol-4-yl]-4-methyl-1,3-thiazol-2-yl]-2,2-dimethyl-propanamide | SMR000156108 | cid_11958611 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H21ClN4O2S2 |
|---|
| Mol. Mass. | 436.979 |
|---|
| SMILES | COc1ccc(Cl)cc1Nc1nc(cs1)-c1sc(NC(=O)C(C)(C)C)nc1C |
|---|
| Structure | 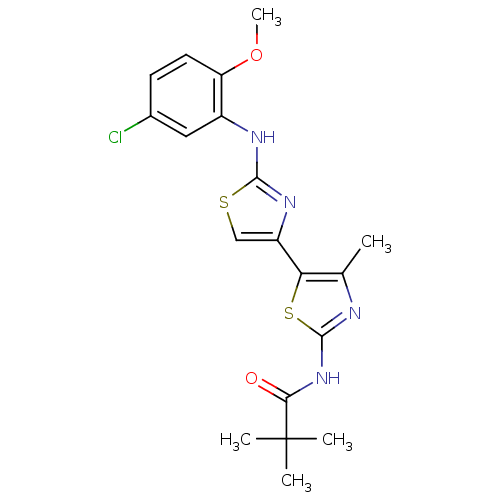
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ye, L; Knapp, JM; Sangwung, P; Fettinger, JC; Verkman, AS; Kurth, MJ Pyrazolylthiazole as DeltaF508-cystic fibrosis transmembrane conductance regulator correctors with improved hydrophilicity compared to bithiazoles. J Med Chem53:3772-81 (2010) [PubMed] Article
Ye, L; Knapp, JM; Sangwung, P; Fettinger, JC; Verkman, AS; Kurth, MJ Pyrazolylthiazole as DeltaF508-cystic fibrosis transmembrane conductance regulator correctors with improved hydrophilicity compared to bithiazoles. J Med Chem53:3772-81 (2010) [PubMed] Article