| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA primase |
|---|
| Ligand | BDBM50034961 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_81442 (CHEMBL690907) |
|---|
| IC50 | 8700.0±n/a nM |
|---|
| Citation |  Crute, JJ; Lehman, IR; Gambino, J; Yang, TF; Medveczky, P; Medveczky, M; Khan, NN; Mulder, C; Monroe, J; Wright, GE Inhibition of herpes simplex virus type 1 helicase-primase by (dichloroanilino)purines and -pyrimidines. J Med Chem38:1820-5 (1995) [PubMed] Crute, JJ; Lehman, IR; Gambino, J; Yang, TF; Medveczky, P; Medveczky, M; Khan, NN; Mulder, C; Monroe, J; Wright, GE Inhibition of herpes simplex virus type 1 helicase-primase by (dichloroanilino)purines and -pyrimidines. J Med Chem38:1820-5 (1995) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| DNA primase |
|---|
| Name: | DNA primase |
|---|
| Synonyms: | DNA helicase/primase complex protein | PRIM_HHV11 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 114429.27 |
|---|
| Organism: | Human herpesvirus 1 (strain 17) (HHV-1) (Human herpes simplex virus1) |
|---|
| Description: | ChEMBL_84513 |
|---|
| Residue: | 1058 |
|---|
| Sequence: | MGQEDGNRGERRAAGTPVEVTALYATDGCVITSSIALLTNSLLGAEPVYIFSYDAYTHDG
RADGPTEQDRFEESRALYQASGGLNGDSFRVTFCLLGTEVGGTHQARGRTRPMFVCRFER
ADDVAALQDALAHGTPLQPDHIAATLDAEATFALHANMILALTVAINNASPRTGRDAAAA
QYDQGASLRSLVGRTSLGQRGLTTLYVHHEVRVLAAYRRAYYGSAQSPFWFLSKFGPDEK
SLVLTTRYYLLQAQRLGGAGATYDLQAIKDICATYAIPHAPRPDTVSAASLTSFAAITRF
CCTSQYARGAAAAGFPLYVERRIAADVRETSALEKFITHDRSCLRVSDREFITYIYLAHF
ECFSPPRLATHLRAVTTHDPNPAASTEQPSPLGREAVEQFFCHVRAQLNIGEYVKHNVTP
RETVLDGDTAKAYLRARTYAPGALTPAPAYCGAVDSATKMMGRLADAEKLLVPRGWPAFA
PASPGEDTAGGTPPPQTCGIVKRLLRLAATEQQGPTPPAIAALIRNAAVQTPLPVYRISM
VPTGQAFAALAWDDWARITRDARLAEAVVSAEAAAHPDHGALGRRLTDRIRAQGPVMPPG
GLDAGGQMYVNRNEIFNGALAITNIILDLDIALKEPVPFRRLHEALGHFRRGALAAVQLL
FPAARVDPDAYPCYFFKSACRPGPASVGSGSGLGNDDDGDWFPCYDDAGDEEWAEDPGAM
DTSHDPPDDEVAYFDLCHEVGPTAEPRETDSPVCSCTDKIGLRVCMPVPAPYVVHGSLTM
RGVARVIQQAVLLDRDFVEAIGSYVKNFLLIDTGVYAHGHSLRLPYFAKIAPDGPACGRL
LPVFVIPPACKDVPAFVAAHADPRRFHFHAPPTYLASPREIRVLHSLGGDYVSFFERKAS
RNALEHFGRRETLTEVLGRYNVQPDAGGTVEGFASELLGRIVACIETHFPEHAGEYQAVS
VRRAVSKDDWVLLQLVPVRGTLQQSLSCLRFKHGRASRATARTFVALSVGANNRLCVSLC
QQCFAAKCDSNRLHTLFTIDAGTPCSPSVPCSTSQPSS
|
|
|
|---|
| BDBM50034961 |
|---|
| n/a |
|---|
| Name | BDBM50034961 |
|---|
| Synonyms: | 6-Chloro-N*4*-(3,4-dichloro-phenyl)-pyrimidine-2,4-diamine | CHEMBL53912 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H7Cl3N4 |
|---|
| Mol. Mass. | 289.548 |
|---|
| SMILES | Nc1nc(Cl)cc(Nc2ccc(Cl)c(Cl)c2)n1 |
|---|
| Structure | 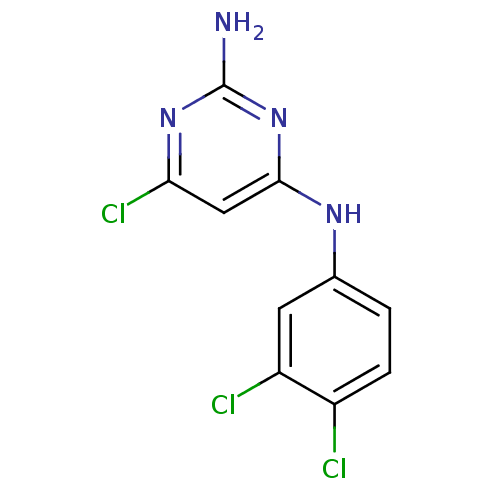
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Crute, JJ; Lehman, IR; Gambino, J; Yang, TF; Medveczky, P; Medveczky, M; Khan, NN; Mulder, C; Monroe, J; Wright, GE Inhibition of herpes simplex virus type 1 helicase-primase by (dichloroanilino)purines and -pyrimidines. J Med Chem38:1820-5 (1995) [PubMed]
Crute, JJ; Lehman, IR; Gambino, J; Yang, TF; Medveczky, P; Medveczky, M; Khan, NN; Mulder, C; Monroe, J; Wright, GE Inhibition of herpes simplex virus type 1 helicase-primase by (dichloroanilino)purines and -pyrimidines. J Med Chem38:1820-5 (1995) [PubMed]