

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Muscarinic acetylcholine receptor M1 | ||
| Ligand | BDBM50010096 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_138134 (CHEMBL747393) | ||
| Ki | 0.051±n/a nM | ||
| Citation |  Nilsson, BM; Sundquist, S; Johansson, G; Nordvall, G; Glas, G; Nilvebrant, L; Hacksell, U 3-Heteroaryl-substituted quinuclidin-3-ol and quinuclidin-2-ene derivatives as muscarinic antagonists. Synthesis and structure-activity relationships. J Med Chem38:473-87 (1995) [PubMed] Nilsson, BM; Sundquist, S; Johansson, G; Nordvall, G; Glas, G; Nilvebrant, L; Hacksell, U 3-Heteroaryl-substituted quinuclidin-3-ol and quinuclidin-2-ene derivatives as muscarinic antagonists. Synthesis and structure-activity relationships. J Med Chem38:473-87 (1995) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Muscarinic acetylcholine receptor M1 | |||
| Name: | Muscarinic acetylcholine receptor M1 | ||
| Synonyms: | ACM1_HUMAN | CHRM1 | Cholinergic receptor, muscarinic 1 | Cholinergic, muscarinic M1 | ||
| Type: | Protein | ||
| Mol. Mass.: | 51442.54 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P11229 | ||
| Residue: | 460 | ||
| Sequence: |
| ||
| BDBM50010096 | |||
| n/a | |||
| Name | BDBM50010096 | ||
| Synonyms: | CHEMBL12980 | Hydroxy-diphenyl-acetic acid 1-aza-bicyclo[2.2.2]oct-3-yl ester | Hydroxy-diphenyl-acetic acid 1-aza-bicyclo[2.2.2]oct-3-yl ester(QNB) | Hydroxy-diphenyl-acetic acid 1-aza-bicyclo[2.2.2]oct-3-yl ester(R)-(-)-(QNB) | ||
| Type | Small organic molecule | ||
| Emp. Form. | C21H23NO3 | ||
| Mol. Mass. | 337.4122 | ||
| SMILES | OC(C(=O)OC1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |(5.41,-4.43,;4.65,-5.78,;6.2,-5.78,;6.97,-7.12,;6.97,-4.45,;8.52,-4.1,;8.52,-2.56,;9.86,-1.78,;11.19,-2.56,;11.19,-4.1,;9.86,-4.88,;9.09,-3.78,;9.09,-2.9,;3.1,-5.81,;2.36,-7.19,;.82,-7.2,;.01,-5.88,;.78,-4.52,;2.32,-4.49,;4.65,-7.23,;3.39,-7.96,;3.38,-9.41,;4.64,-10.15,;5.91,-9.42,;5.91,-7.97,)| | ||
| Structure | 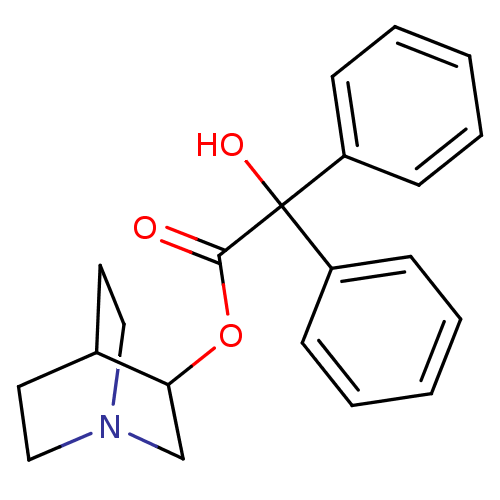
| ||