| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Purine nucleoside phosphorylase |
|---|
| Ligand | BDBM50039547 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_162034 (CHEMBL766674) |
|---|
| IC50 | 51±n/a nM |
|---|
| Citation |  Niwas, S; Chand, P; Pathak, VP; Montgomery, JA Structure-based design of inhibitors of purine nucleoside phosphorylase. 5. 9-Deazahypoxanthines. J Med Chem37:2477-80 (1994) [PubMed] Niwas, S; Chand, P; Pathak, VP; Montgomery, JA Structure-based design of inhibitors of purine nucleoside phosphorylase. 5. 9-Deazahypoxanthines. J Med Chem37:2477-80 (1994) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Purine nucleoside phosphorylase |
|---|
| Name: | Purine nucleoside phosphorylase |
|---|
| Synonyms: | Inosine phosphorylase | Np | PNPH_RAT | Pnp | Purine nucleoside phosphorylase | Purine-nucleoside phosphorylase | nucleoside phosphorylase |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 32303.61 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_162034 |
|---|
| Residue: | 289 |
|---|
| Sequence: | MENEFTYEDYQRTAEWLRSHTKHRPQVAVICGSGLGGLTAKLTQPQAFDYNEIPNFPQST
VQGHAGRLVFGFLNGRSCVMMQGRFHMYEGYSLSKVTFPVRVFHLLGVDTLVVTNAAGGL
NPKFEVGDIMLIRDHINLPGFCGQNPLRGPNDERFGVRFPAMSDAYDRDMRQKAFNAWKQ
MGEQRELQEGTYIMSAGPTFETVAESCLLRMLGADAVGMSTVPEVIVARHCGLRVFGFSL
ITNKVVMDYNNLEKASHQEVLEAGKAAAQKLEQFVSILMESIPPRERAN
|
|
|
|---|
| BDBM50039547 |
|---|
| n/a |
|---|
| Name | BDBM50039547 |
|---|
| Synonyms: | 2-Amino-7-benzyl-3,5-dihydro-pyrrolo[3,2-d]pyrimidin-4-one | 2-amino-7-benzyl-3H-pyrrolo[3,2-d]pyrimidin-4(5H)-one | CHEMBL310202 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H12N4O |
|---|
| Mol. Mass. | 240.2606 |
|---|
| SMILES | Nc1nc2c(Cc3ccccc3)c[nH]c2c(=O)[nH]1 |
|---|
| Structure | 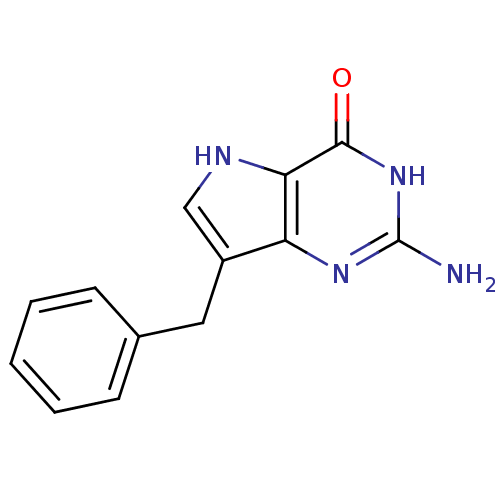
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Niwas, S; Chand, P; Pathak, VP; Montgomery, JA Structure-based design of inhibitors of purine nucleoside phosphorylase. 5. 9-Deazahypoxanthines. J Med Chem37:2477-80 (1994) [PubMed]
Niwas, S; Chand, P; Pathak, VP; Montgomery, JA Structure-based design of inhibitors of purine nucleoside phosphorylase. 5. 9-Deazahypoxanthines. J Med Chem37:2477-80 (1994) [PubMed]