| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Purine nucleoside phosphorylase |
|---|
| Ligand | BDBM50039552 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_162190 (CHEMBL766710) |
|---|
| Ki | 210±n/a nM |
|---|
| Citation |  Guida, WC; Elliott, RD; Thomas, HJ; Secrist, JA; Babu, YS; Bugg, CE; Erion, MD; Ealick, SE; Montgomery, JA Structure-based design of inhibitors of purine nucleoside phosphorylase. 4. A study of phosphate mimics. J Med Chem37:1109-14 (1994) [PubMed] Guida, WC; Elliott, RD; Thomas, HJ; Secrist, JA; Babu, YS; Bugg, CE; Erion, MD; Ealick, SE; Montgomery, JA Structure-based design of inhibitors of purine nucleoside phosphorylase. 4. A study of phosphate mimics. J Med Chem37:1109-14 (1994) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Purine nucleoside phosphorylase |
|---|
| Name: | Purine nucleoside phosphorylase |
|---|
| Synonyms: | Inosine phosphorylase | Inosine-guanosine phosphorylase | NP | PNP | PNPH_HUMAN | Purine nucleoside phosphorylase (PNPase) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 32119.53 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 289 |
|---|
| Sequence: | MENGYTYEDYKNTAEWLLSHTKHRPQVAIICGSGLGGLTDKLTQAQIFDYGEIPNFPRST
VPGHAGRLVFGFLNGRACVMMQGRFHMYEGYPLWKVTFPVRVFHLLGVDTLVVTNAAGGL
NPKFEVGDIMLIRDHINLPGFSGQNPLRGPNDERFGDRFPAMSDAYDRTMRQRALSTWKQ
MGEQRELQEGTYVMVAGPSFETVAECRVLQKLGADAVGMSTVPEVIVARHCGLRVFGFSL
ITNKVIMDYESLEKANHEEVLAAGKQAAQKLEQFVSILMASIPLPDKAS
|
|
|
|---|
| BDBM50039552 |
|---|
| n/a |
|---|
| Name | BDBM50039552 |
|---|
| Synonyms: | CHEMBL267312 | [3,3-Dimethyl-5-(6-oxo-1,6-dihydro-purin-9-yl)-pentyl]-phosphonic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H19N4O4P |
|---|
| Mol. Mass. | 314.2774 |
|---|
| SMILES | CC(C)(CCn1cnc2c1nc[nH]c2=O)CCP(O)(O)=O |
|---|
| Structure | 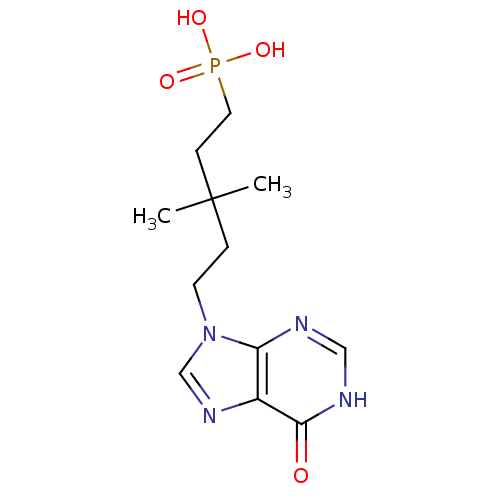
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Guida, WC; Elliott, RD; Thomas, HJ; Secrist, JA; Babu, YS; Bugg, CE; Erion, MD; Ealick, SE; Montgomery, JA Structure-based design of inhibitors of purine nucleoside phosphorylase. 4. A study of phosphate mimics. J Med Chem37:1109-14 (1994) [PubMed]
Guida, WC; Elliott, RD; Thomas, HJ; Secrist, JA; Babu, YS; Bugg, CE; Erion, MD; Ealick, SE; Montgomery, JA Structure-based design of inhibitors of purine nucleoside phosphorylase. 4. A study of phosphate mimics. J Med Chem37:1109-14 (1994) [PubMed]