| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Gag-Pol polyprotein [489-587] |
|---|
| Ligand | BDBM905 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_159974 (CHEMBL768121) |
|---|
| Ki | 43±n/a nM |
|---|
| Citation |  Reddy, MR; Varney, MD; Kalish, V; Viswanadhan, VN; Appelt, K Calculation of relative differences in the binding free energies of HIV1 protease inhibitors: a thermodynamic cycle perturbation approach. J Med Chem37:1145-52 (1994) [PubMed] Reddy, MR; Varney, MD; Kalish, V; Viswanadhan, VN; Appelt, K Calculation of relative differences in the binding free energies of HIV1 protease inhibitors: a thermodynamic cycle perturbation approach. J Med Chem37:1145-52 (1994) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Gag-Pol polyprotein [489-587] |
|---|
| Name: | Gag-Pol polyprotein [489-587] |
|---|
| Synonyms: | Human immunodeficiency virus type 1 protease | POL_HV1H2 | Pol polyprotein | gag-pol |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 10781.16 |
|---|
| Organism: | Human immunodeficiency virus type 1 |
|---|
| Description: | P04585[489-587] |
|---|
| Residue: | 99 |
|---|
| Sequence: | PQVTLWQRPLVTIKIGGQLKEALLDTGADDTVLEEMSLPGRWKPKMIGGIGGFIKVRQYD
QILIEICGHKAIGTVLVGPTPVNIIGRNLLTQIGCTLNF
|
|
|
|---|
| BDBM905 |
|---|
| n/a |
|---|
| Name | BDBM905 |
|---|
| Synonyms: | (4S,5S)-5-[(2S)-2-[(2S)-2-aminopropanamido]propanamido]-4-hydroxy-N-[1H-indol-2-yl(phenyl)methyl]-6-phenylhexanamide | 5-(L-Alanyl-L-alanylamino)-4-hydroxy-6-phenylhexanoic Acid ((1H-Indol-2-y1)phenylmethyl)amide | C-terminal inhibitor 3 | CHEMBL14972 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C33H39N5O4 |
|---|
| Mol. Mass. | 569.6939 |
|---|
| SMILES | C[C@H](N)C(=O)N[C@@H](C)C(=O)N[C@@H](Cc1ccccc1)[C@@H](O)CCC(=O)NC(c1cc2ccccc2[nH]1)c1ccccc1 |r| |
|---|
| Structure | 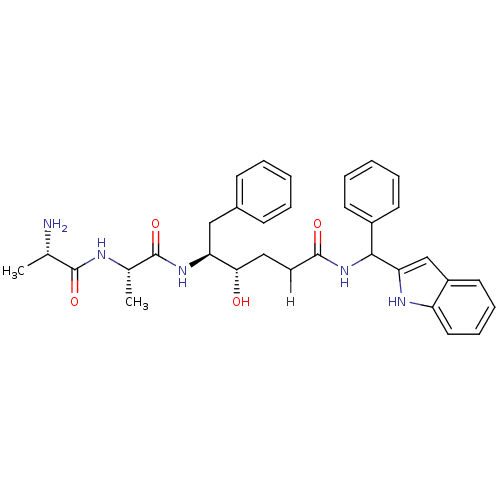
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Reddy, MR; Varney, MD; Kalish, V; Viswanadhan, VN; Appelt, K Calculation of relative differences in the binding free energies of HIV1 protease inhibitors: a thermodynamic cycle perturbation approach. J Med Chem37:1145-52 (1994) [PubMed]
Reddy, MR; Varney, MD; Kalish, V; Viswanadhan, VN; Appelt, K Calculation of relative differences in the binding free energies of HIV1 protease inhibitors: a thermodynamic cycle perturbation approach. J Med Chem37:1145-52 (1994) [PubMed]