

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Ligand | BDBM50500429 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1546366 (CHEMBL3748597) | ||
| Ki | 112±n/a nM | ||
| Citation |  Ouach, A; Pin, F; Bertrand, E; Vercouillie, J; Gulhan, Z; Mothes, C; Deloye, JB; Guilloteau, D; Suzenet, F; Chalon, S; Routier, S Design of ?7 nicotinic acetylcholine receptor ligands using the (het)Aryl-1,2,3-triazole core: Synthesis, in vitro evaluation and SAR studies. Eur J Med Chem107:153-64 (2016) [PubMed] Article Ouach, A; Pin, F; Bertrand, E; Vercouillie, J; Gulhan, Z; Mothes, C; Deloye, JB; Guilloteau, D; Suzenet, F; Chalon, S; Routier, S Design of ?7 nicotinic acetylcholine receptor ligands using the (het)Aryl-1,2,3-triazole core: Synthesis, in vitro evaluation and SAR studies. Eur J Med Chem107:153-64 (2016) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neuronal acetylcholine receptor subunit alpha-7 | |||
| Name: | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Synonyms: | ACHA7_RAT | Acra7 | Cholinergic, Nicotinic Alpha7 | Cholinergic, Nicotinic Alpha7/5-HT3 | Chrna7 | Neuronal acetylcholine receptor | Neuronal acetylcholine receptor (alpha7 nAChR) | Neuronal acetylcholine receptor subunit alpha 7 | Neuronal acetylcholine receptor subunit alpha-7 | Neuronal acetylcholine receptor subunit alpha-7 (nAChR alpha7) | Neuronal acetylcholine receptor subunit alpha-7 (nAChR) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 56502.44 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | Q05941 | ||
| Residue: | 502 | ||
| Sequence: |
| ||
| BDBM50500429 | |||
| n/a | |||
| Name | BDBM50500429 | ||
| Synonyms: | CHEMBL3745951 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C16H20N4O | ||
| Mol. Mass. | 284.3562 | ||
| SMILES | COc1ccccc1-c1cn(nn1)[C@H]1CN2CCC1CC2 |r,wD:13.14,(1.81,3.95,;1.15,2.91,;-.39,2.97,;-1.11,4.33,;-2.65,4.39,;-3.47,3.08,;-2.75,1.72,;-1.2,1.67,;-.59,.25,;.92,-.08,;1.05,-1.6,;-.35,-2.23,;-1.37,-1.07,;2.43,-2.41,;2.41,-4.03,;3.81,-4.83,;5.21,-4.01,;5.21,-2.38,;3.81,-1.57,;3.26,-2.74,;4.33,-3.38,)| | ||
| Structure | 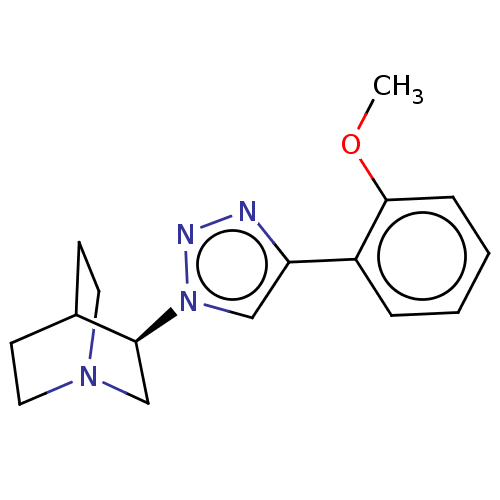
| ||