| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Protein mono-ADP-ribosyltransferase PARP12 |
|---|
| Ligand | BDBM50503853 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1812389 (CHEMBL4311849) |
|---|
| IC50 | >30000±n/a nM |
|---|
| Citation |  Morgan, RK; Kirby, IT; Vermehren-Schmaedick, A; Rodriguez, K; Cohen, MS Rational Design of Cell-Active Inhibitors of PARP10. ACS Med Chem Lett10:74-79 (2019) [PubMed] Article Morgan, RK; Kirby, IT; Vermehren-Schmaedick, A; Rodriguez, K; Cohen, MS Rational Design of Cell-Active Inhibitors of PARP10. ACS Med Chem Lett10:74-79 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Protein mono-ADP-ribosyltransferase PARP12 |
|---|
| Name: | Protein mono-ADP-ribosyltransferase PARP12 |
|---|
| Synonyms: | ADP-ribosyltransferase diphtheria toxin-like 12 | ARTD12 | PAR12_HUMAN | PARP-12 | PARP12 | Poly [ADP-ribose] polymerase 12 | ZC3HDC1 | Zinc finger CCCH domain-containing protein 1 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 79092.00 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_107665 |
|---|
| Residue: | 701 |
|---|
| Sequence: | MAQAGVVGEVTQVLCAAGGALELPELRRRLRMGLSADALERLLRQRGRFVVAVRAGGAAA
APERVVLAASPLRLCRAHQGSKPGCVGLCAQLHLCRFMVYGACKFLRAGKNCRNSHSLTT
EHNLSVLRTHGVDHLSYNELCQLLFQNDPWLLPEICQHYNKGDGPHGSCAFQKQCIKLHI
CQYFLQGECKFGTSCKRSHDFSNSENLEKLEKLGMSSDLVSRLPTIYRNAHDIKNKSSAP
SRVPPLFVPQGTSERKDSSGSVSPNTLSQEEGDQICLYHIRKSCSFQDKCHRVHFHLPYR
WQFLDRGKWEDLDNMELIEEAYCNPKIERILCSESASTFHSHCLNFNAMTYGATQARRLS
TASSVTKPPHFILTTDWIWYWSDEFGSWQEYGRQGTVHPVTTVSSSDVEKAYLAYCTPGS
DGQAATLKFQAGKHNYELDFKAFVQKNLVYGTTKKVCRRPKYVSPQDVTTMQTCNTKFPG
PKSIPDYWDSSALPDPGFQKITLSSSSEEYQKVWNLFNRTLPFYFVQKIERVQNLALWEV
YQWQKGQMQKQNGGKAVDERQLFHGTSAIFVDAICQQNFDWRVCGVHGTSYGKGSYFARD
AAYSHHYSKSDTQTHTMFLARVLVGEFVRGNASFVRPPAKEGWSNAFYDSCVNSVSDPSI
FVIFEKHQVYPEYVIQYTTSSKPSVTPSILLALGSLFSSRQ
|
|
|
|---|
| BDBM50503853 |
|---|
| n/a |
|---|
| Name | BDBM50503853 |
|---|
| Synonyms: | CHEMBL4545565 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H13F3N2O |
|---|
| Mol. Mass. | 306.2824 |
|---|
| SMILES | Cc1c2CCNC(=O)c2ccc1-c1ccnc(c1)C(F)(F)F |
|---|
| Structure | 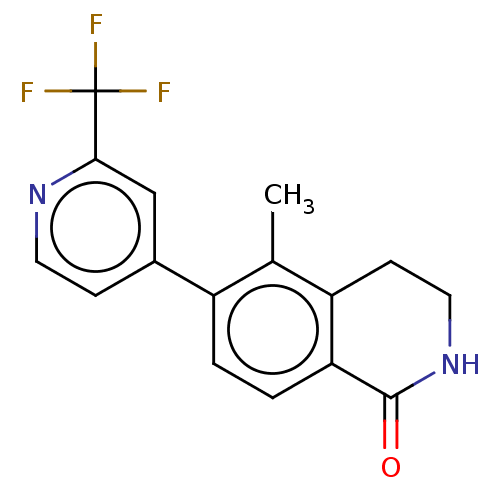
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Morgan, RK; Kirby, IT; Vermehren-Schmaedick, A; Rodriguez, K; Cohen, MS Rational Design of Cell-Active Inhibitors of PARP10. ACS Med Chem Lett10:74-79 (2019) [PubMed] Article
Morgan, RK; Kirby, IT; Vermehren-Schmaedick, A; Rodriguez, K; Cohen, MS Rational Design of Cell-Active Inhibitors of PARP10. ACS Med Chem Lett10:74-79 (2019) [PubMed] Article