| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 10 |
|---|
| Ligand | BDBM50506712 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1827111 (CHEMBL4326985) |
|---|
| Kd | 710±n/a nM |
|---|
| Citation |  Schepetkin, IA; Khlebnikov, AI; Potapov, AS; Kovrizhina, AR; Matveevskaya, VV; Belyanin, ML; Atochin, DN; Zanoza, SO; Gaidarzhy, NM; Lyakhov, SA; Kirpotina, LN; Quinn, MT Synthesis, biological evaluation, and molecular modeling of 11H-indeno[1,2-b]quinoxalin-11-one derivatives and tryptanthrin-6-oxime as c-Jun N-terminal kinase inhibitors. Eur J Med Chem161:179-191 (2019) [PubMed] Article Schepetkin, IA; Khlebnikov, AI; Potapov, AS; Kovrizhina, AR; Matveevskaya, VV; Belyanin, ML; Atochin, DN; Zanoza, SO; Gaidarzhy, NM; Lyakhov, SA; Kirpotina, LN; Quinn, MT Synthesis, biological evaluation, and molecular modeling of 11H-indeno[1,2-b]quinoxalin-11-one derivatives and tryptanthrin-6-oxime as c-Jun N-terminal kinase inhibitors. Eur J Med Chem161:179-191 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 10 |
|---|
| Name: | Mitogen-activated protein kinase 10 |
|---|
| Synonyms: | JNK3 | JNK3A | MAP kinase p49 3F12 | MAPK10 | MK10_HUMAN | Mitogen-Activated Protein Kinase 10 (JNK3) | Mitogen-activated protein kinase 10 (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12) | Mitogen-activated protein kinase 10/Receptor-interacting serine/threonine-protein kinase 1 | PRKM10 | SAPK1B | Stress-activated protein kinase JNK3 | c-Jun N-terminal kinase 3 (JNK3) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52586.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 464 |
|---|
| Sequence: | MSLHFLYYCSEPTLDVKIAFCQGFDKQVDVSYIAKHYNMSKSKVDNQFYSVEVGDSTFTV
LKRYQNLKPIGSGAQGIVCAAYDAVLDRNVAIKKLSRPFQNQTHAKRAYRELVLMKCVNH
KNIISLLNVFTPQKTLEEFQDVYLVMELMDANLCQVIQMELDHERMSYLLYQMLCGIKHL
HSAGIIHRDLKPSNIVVKSDCTLKILDFGLARTAGTSFMMTPYVVTRYYRAPEVILGMGY
KENVDIWSVGCIMGEMVRHKILFPGRDYIDQWNKVIEQLGTPCPEFMKKLQPTVRNYVEN
RPKYAGLTFPKLFPDSLFPADSEHNKLKASQARDLLSKMLVIDPAKRISVDDALQHPYIN
VWYDPAEVEAPPPQIYDKQLDEREHTIEEWKELIYKEVMNSEEKTKNGVVKGQPSPSGAA
VNSSESLPPSSSVNDISSMSTDQTLASDTDSSLEASAGPLGCCR
|
|
|
|---|
| BDBM50506712 |
|---|
| n/a |
|---|
| Name | BDBM50506712 |
|---|
| Synonyms: | CHEMBL4583031 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H8ClN3O |
|---|
| Mol. Mass. | 281.697 |
|---|
| SMILES | O\N=C1\c2ccccc2-c2nc3cc(Cl)ccc3nc12 |
|---|
| Structure | 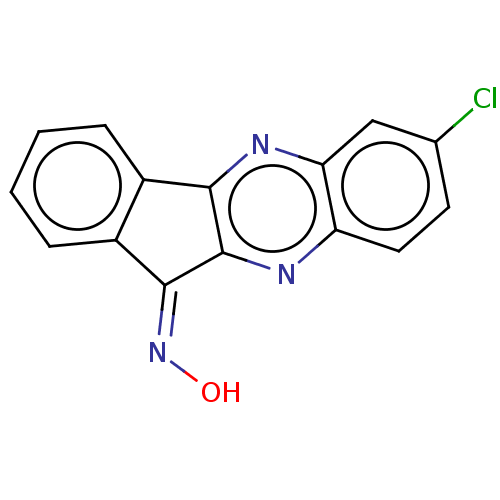
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Schepetkin, IA; Khlebnikov, AI; Potapov, AS; Kovrizhina, AR; Matveevskaya, VV; Belyanin, ML; Atochin, DN; Zanoza, SO; Gaidarzhy, NM; Lyakhov, SA; Kirpotina, LN; Quinn, MT Synthesis, biological evaluation, and molecular modeling of 11H-indeno[1,2-b]quinoxalin-11-one derivatives and tryptanthrin-6-oxime as c-Jun N-terminal kinase inhibitors. Eur J Med Chem161:179-191 (2019) [PubMed] Article
Schepetkin, IA; Khlebnikov, AI; Potapov, AS; Kovrizhina, AR; Matveevskaya, VV; Belyanin, ML; Atochin, DN; Zanoza, SO; Gaidarzhy, NM; Lyakhov, SA; Kirpotina, LN; Quinn, MT Synthesis, biological evaluation, and molecular modeling of 11H-indeno[1,2-b]quinoxalin-11-one derivatives and tryptanthrin-6-oxime as c-Jun N-terminal kinase inhibitors. Eur J Med Chem161:179-191 (2019) [PubMed] Article