| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cGMP-dependent 3',5'-cyclic phosphodiesterase |
|---|
| Ligand | BDBM50061899 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_156302 (CHEMBL760823) |
|---|
| IC50 | 11000±n/a nM |
|---|
| Citation |  Xia, Y; Chackalamannil, S; Czarniecki, M; Tsai, H; Vaccaro, H; Cleven, R; Cook, J; Fawzi, A; Watkins, R; Zhang, H Synthesis and evaluation of polycyclic pyrazolo[3,4-d]pyrimidines as PDE1 and PDE5 cGMP phosphodiesterase inhibitors. J Med Chem40:4372-7 (1998) [PubMed] Article Xia, Y; Chackalamannil, S; Czarniecki, M; Tsai, H; Vaccaro, H; Cleven, R; Cook, J; Fawzi, A; Watkins, R; Zhang, H Synthesis and evaluation of polycyclic pyrazolo[3,4-d]pyrimidines as PDE1 and PDE5 cGMP phosphodiesterase inhibitors. J Med Chem40:4372-7 (1998) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cGMP-dependent 3',5'-cyclic phosphodiesterase |
|---|
| Name: | cGMP-dependent 3',5'-cyclic phosphodiesterase |
|---|
| Synonyms: | PDE2A | PDE2A_BOVIN | Phosphodiesterase 2A |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 103200.18 |
|---|
| Organism: | Bos taurus |
|---|
| Description: | ChEMBL_154420 |
|---|
| Residue: | 921 |
|---|
| Sequence: | MRRQPAASRDLFAQEPVPPGSGDGALQDALLSLGSVIDVAGLQQAVKEALSAVLPKVETV
YTYLLDGESRLVCEEPPHELPQEGKVREAVISRKRLGCNGLGPSDLPGKPLARLVAPLAP
DTQVLVIPLVDKEAGAVAAVILVHCGQLSDNEEWSLQAVEKHTLVALKRVQALQQRESSV
APEATQNPPEEAAGDQKGGVAYTNQDRKILQLCGELYDLDASSLQLKVLQYLQQETQASR
CCLLLVSEDNLQLSCKVIGDKVLEEEISFPLTTGRLGQVVEDKKSIQLKDLTSEDMQQLQ
SMLGCEVQAMLCVPVISRATDQVVALACAFNKLGGDLFTDQDEHVIQHCFHYTSTVLTST
LAFQKEQKLKCECQALLQVAKNLFTHLDDVSVLLQEIITEARNLSNAEICSVFLLDQNEL
VAKVFDGGVVEDESYEIRIPADQGIAGHVATTGQILNIPDAYAHPLFYRGVDDSTGFRTR
NILCFPIKNENQEVIGVAELVNKINGPWFSKFDEDLATAFSIYCGISIAHSLLYKKVNEA
QYRSHLANEMMMYHMKVSDDEYTKLLHDGIQPVAAIDSNFASFTYTPRSLPEDDTSMAIL
SMLQDMNFINNYKIDCPTLARFCLMVKKGYRDPPYHNWMHAFSVSHFCYLLYKNLELTNY
LEDMEIFALFISCMCHDLDHRGTNNSFQVASKSVLAALYSSEGSVMERHHFAQAIAILNT
HGCNIFDHFSRKDYQRMLDLMRDIILATDLAHHLRIFKDLQKMAEVGYDRTNKQHHSLLL
CLLMTSCDLSDQTKGWKTTRKIAELIYKEFFSQGDLEKAMGNRPMEMMDREKAYIPELQI
SFMEHIAMPIYKLLQDLFPKAAELYERVASNREHWTKVSHKFTIRGLPSNNSLDFLDEEY
EVPDLDGARAPINGCCSLDAE
|
|
|
|---|
| BDBM50061899 |
|---|
| n/a |
|---|
| Name | BDBM50061899 |
|---|
| Synonyms: | 2',5'-Dimethyl-3'-(phenylmethyl)spiro[cyclopentane1,7'(8'H)-[2H]imidazol[1,2-a]pyrzolo[4,3-e]pyrimidin]-4'-(5'H)-one | CHEMBL137400 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H23N5O |
|---|
| Mol. Mass. | 349.4295 |
|---|
| SMILES | CN1C2=NC3(CCCC3)CN2c2nn(C)c(Cc3ccccc3)c2C1=O |t:2| |
|---|
| Structure | 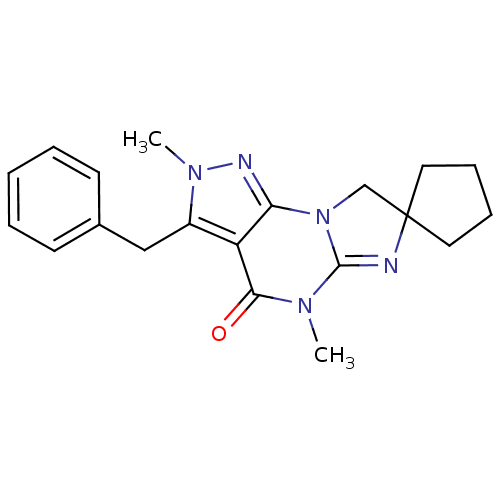
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Xia, Y; Chackalamannil, S; Czarniecki, M; Tsai, H; Vaccaro, H; Cleven, R; Cook, J; Fawzi, A; Watkins, R; Zhang, H Synthesis and evaluation of polycyclic pyrazolo[3,4-d]pyrimidines as PDE1 and PDE5 cGMP phosphodiesterase inhibitors. J Med Chem40:4372-7 (1998) [PubMed] Article
Xia, Y; Chackalamannil, S; Czarniecki, M; Tsai, H; Vaccaro, H; Cleven, R; Cook, J; Fawzi, A; Watkins, R; Zhang, H Synthesis and evaluation of polycyclic pyrazolo[3,4-d]pyrimidines as PDE1 and PDE5 cGMP phosphodiesterase inhibitors. J Med Chem40:4372-7 (1998) [PubMed] Article