

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mitogen-activated protein kinase 14 | ||
| Ligand | BDBM50313107 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1837226 (CHEMBL4337359) | ||
| Ki | 1.000000±n/a nM | ||
| Citation |  Astolfi, A; Kudolo, M; Brea, J; Manni, G; Manfroni, G; Palazzotti, D; Sabatini, S; Cecchetti, F; Felicetti, T; Cannalire, R; Massari, S; Tabarrini, O; Loza, MI; Fallarino, F; Cecchetti, V; Laufer, SA; Barreca, ML Discovery of potent p38? MAPK inhibitors through a funnel like workflow combining in silico screening and in vitro validation. Eur J Med Chem182:0 (2019) [PubMed] Article Astolfi, A; Kudolo, M; Brea, J; Manni, G; Manfroni, G; Palazzotti, D; Sabatini, S; Cecchetti, F; Felicetti, T; Cannalire, R; Massari, S; Tabarrini, O; Loza, MI; Fallarino, F; Cecchetti, V; Laufer, SA; Barreca, ML Discovery of potent p38? MAPK inhibitors through a funnel like workflow combining in silico screening and in vitro validation. Eur J Med Chem182:0 (2019) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mitogen-activated protein kinase 14 | |||
| Name: | Mitogen-activated protein kinase 14 | ||
| Synonyms: | CSAID-binding protein | CSBP | CSBP1 | CSBP2 | CSPB1 | Cytokine suppressive anti-inflammatory drug-binding protein | MAP kinase 14 | MAP kinase MXI2 | MAP kinase p38 alpha | MAPK 14 | MAPK14 | MAX-interacting protein 2 | MK14_HUMAN | MXI2 | Mitogen-activated protein kinase p38 alpha | SAPK2A | Stress-activated protein kinase 2a | p38 MAP kinase alpha/beta | ||
| Type: | Serine/threonine-protein kinase | ||
| Mol. Mass.: | 41286.76 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q16539 | ||
| Residue: | 360 | ||
| Sequence: |
| ||
| BDBM50313107 | |||
| n/a | |||
| Name | BDBM50313107 | ||
| Synonyms: | 4-chloro-N-cyclopropyl-3-(1-(2,6-difluorophenyl)-1H-pyrazolo[4,3-d]pyridazin-4-ylamino)benzamide | 4-chloro-N-cyclopropyl-3-{[1-(2,6-difluorophenyl)-1H-pyrazolo[3,4-d]pyridazin-4-yl]amino}benzamide | CHEMBL1082158 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C21H15ClF2N6O | ||
| Mol. Mass. | 440.833 | ||
| SMILES | Fc1cccc(F)c1-n1ncc2c(Nc3cc(ccc3Cl)C(=O)NC3CC3)nncc12 |(-6.01,-8.16,;-5.23,-9.48,;-5.98,-10.82,;-5.21,-12.15,;-3.66,-12.13,;-2.9,-10.79,;-1.37,-10.77,;-3.69,-9.47,;-2.93,-8.14,;-3.84,-6.9,;-2.95,-5.64,;-1.48,-6.1,;-.16,-5.33,;-.1,-3.79,;1.27,-3.07,;2.56,-3.9,;3.92,-3.18,;3.98,-1.64,;2.68,-.83,;1.32,-1.54,;.02,-.73,;5.22,-4.01,;5.16,-5.55,;6.59,-3.29,;7.89,-4.12,;8.6,-5.48,;9.43,-4.18,;1.18,-6.08,;1.19,-7.63,;-.14,-8.4,;-1.47,-7.64,)| | ||
| Structure | 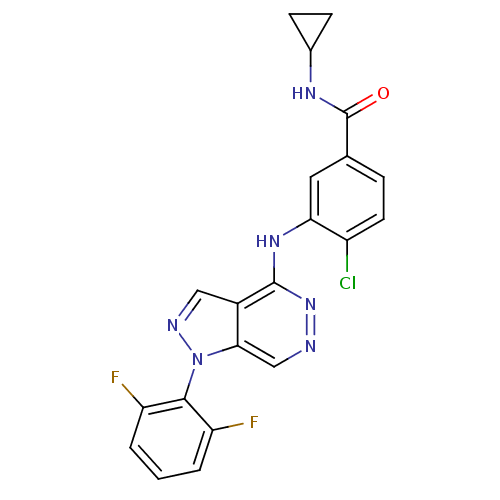
| ||