| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Genome polyprotein |
|---|
| Ligand | BDBM50075610 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_161099 (CHEMBL771560) |
|---|
| EC50 | 10±n/a nM |
|---|
| Citation |  Dragovich, PS; Prins, TJ; Zhou, R; Webber, SE; Marakovits, JT; Fuhrman, SA; Patick, AK; Matthews, DA; Lee, CA; Ford, CE; Burke, BJ; Rejto, PA; Hendrickson, TF; Tuntland, T; Brown, EL; Meador, JW; Ferre, RA; Harr, JE; Kosa, MB; Worland, ST Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as L-glutamine replacements. J Med Chem42:1213-24 (1999) [PubMed] Article Dragovich, PS; Prins, TJ; Zhou, R; Webber, SE; Marakovits, JT; Fuhrman, SA; Patick, AK; Matthews, DA; Lee, CA; Ford, CE; Burke, BJ; Rejto, PA; Hendrickson, TF; Tuntland, T; Brown, EL; Meador, JW; Ferre, RA; Harr, JE; Kosa, MB; Worland, ST Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as L-glutamine replacements. J Med Chem42:1213-24 (1999) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Genome polyprotein |
|---|
| Name: | Genome polyprotein |
|---|
| Synonyms: | Human rhinovirus A protease | Human rhinovirus B 3A protease |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 44361.04 |
|---|
| Organism: | Human rhinovirus B |
|---|
| Description: | ChEMBL_158953 |
|---|
| Residue: | 401 |
|---|
| Sequence: | AFRPCNVNTKIGNAKCCPFVCGKAVTFKDRSTCSTYNLSSSLHHILEEDKRRRQVVDVMS
AIFQGPISLDAPPPPAIADLLQSVRTPRVIKYCQIIMGHPAECQVERDLNIANSIIAIIA
NIISIAGIIFVIYKLFCSLQGPYSGEPKPKTKVPERRVVAQGPEEEFGRSILKNNTCVIT
TGNGKFTGLGIHDRILIIPTHADPGREVQVNGVHTKVLDSYDLYNRDGVKLEITVIQLDR
NEKFRDIRKYIPETEDDYPECNLALSANQDEPTIIKVGDVVSYGNILLSGNQTARMLKYN
YPTKSGYCGGVLYKIGQILGIHVGGNGRDGFSAMLLRSYFTGQIKVNKHATECGLPDIQT
IHTPSKTKLQPSVFYDVFPGSKEPAVLTDNDPRLEVNFKEA
|
|
|
|---|
| BDBM50075610 |
|---|
| n/a |
|---|
| Name | BDBM50075610 |
|---|
| Synonyms: | (E)-(S)-4-[(S)-2-{(S)-3,3-Dimethyl-2-[(5-methyl-isoxazole-3-carbonyl)-amino]-butyrylamino}-3-(4-fluoro-phenyl)-propionylamino]-5-((S)-2-oxo-pyrrolidin-3-yl)-pent-2-enoic acid ethyl ester | CHEMBL21082 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H40FN5O7 |
|---|
| Mol. Mass. | 613.677 |
|---|
| SMILES | CCOC(=O)\C=C\[C@H](C[C@@H]1CCNC1=O)NC(=O)[C@H](Cc1ccc(F)cc1)NC(=O)[C@@H](NC(=O)c1cc(C)on1)C(C)(C)C |
|---|
| Structure | 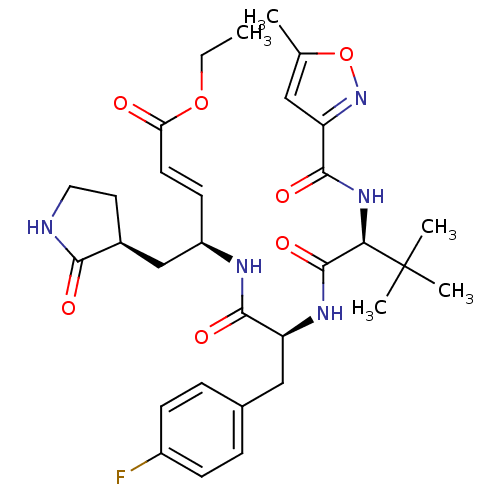
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Dragovich, PS; Prins, TJ; Zhou, R; Webber, SE; Marakovits, JT; Fuhrman, SA; Patick, AK; Matthews, DA; Lee, CA; Ford, CE; Burke, BJ; Rejto, PA; Hendrickson, TF; Tuntland, T; Brown, EL; Meador, JW; Ferre, RA; Harr, JE; Kosa, MB; Worland, ST Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as L-glutamine replacements. J Med Chem42:1213-24 (1999) [PubMed] Article
Dragovich, PS; Prins, TJ; Zhou, R; Webber, SE; Marakovits, JT; Fuhrman, SA; Patick, AK; Matthews, DA; Lee, CA; Ford, CE; Burke, BJ; Rejto, PA; Hendrickson, TF; Tuntland, T; Brown, EL; Meador, JW; Ferre, RA; Harr, JE; Kosa, MB; Worland, ST Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as L-glutamine replacements. J Med Chem42:1213-24 (1999) [PubMed] Article