

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Delta-type opioid receptor | ||
| Ligand | BDBM50344435 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1884534 (CHEMBL4386116) | ||
| Ki | 371±n/a nM | ||
| Citation |  Kronenberg, E; Weber, F; Brune, S; Schepmann, D; Almansa, C; Friedland, K; Laurini, E; Pricl, S; WŁnsch, B Synthesis and Structure-Affinity Relationships of Spirocyclic Benzopyrans with Exocyclic Amino Moiety. J Med Chem62:4204-4217 (2019) [PubMed] Article Kronenberg, E; Weber, F; Brune, S; Schepmann, D; Almansa, C; Friedland, K; Laurini, E; Pricl, S; WŁnsch, B Synthesis and Structure-Affinity Relationships of Spirocyclic Benzopyrans with Exocyclic Amino Moiety. J Med Chem62:4204-4217 (2019) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Delta-type opioid receptor | |||
| Name: | Delta-type opioid receptor | ||
| Synonyms: | Cytochrome P450 3A4 | DOR-1 | Delta opioid receptor | Delta-type opioid receptor | Delta-type opioid receptor (DOR) | OPIATE Delta | OPRD_RAT | Opiate Delta 1 | Opioid receptor | Opioid receptor A | Opioid receptors; mu & delta | Oprd1 | Ror-a | Voltage-gated potassium channel | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 40465.04 | ||
| Organism: | Rattus norvegicus (rat) | ||
| Description: | Competition binding assays were using CHO-K1 cell membranes expressing the opioid receptor. | ||
| Residue: | 372 | ||
| Sequence: |
| ||
| BDBM50344435 | |||
| n/a | |||
| Name | BDBM50344435 | ||
| Synonyms: | CHEMBL1779999 | trans-N-Benzyl-3-methoxy-N-methyl-3,4-dihydrospiro-[[2]benzopyran-1,1'-cyclohexan]-4'-amine | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H29NO2 | ||
| Mol. Mass. | 351.4819 | ||
| SMILES | COC1Cc2ccccc2[C@@]2(CC[C@@H](CC2)N(C)Cc2ccccc2)O1 |r,wU:10.27,wD:13.17,(-1.47,-19.23,;-2.8,-20,;-2.81,-21.55,;-1.48,-22.32,;-1.48,-23.86,;-.17,-24.62,;-.17,-26.14,;-1.5,-26.91,;-2.81,-26.14,;-2.81,-24.62,;-4.14,-23.85,;-4.91,-25.2,;-6.63,-24.58,;-7.82,-25,;-7.05,-23.64,;-5.41,-24.27,;-7.83,-26.55,;-6.49,-27.32,;-9.17,-27.31,;-10.5,-26.53,;-10.49,-24.99,;-11.83,-24.21,;-13.17,-24.98,;-13.17,-26.53,;-11.84,-27.3,;-4.15,-22.31,)| | ||
| Structure | 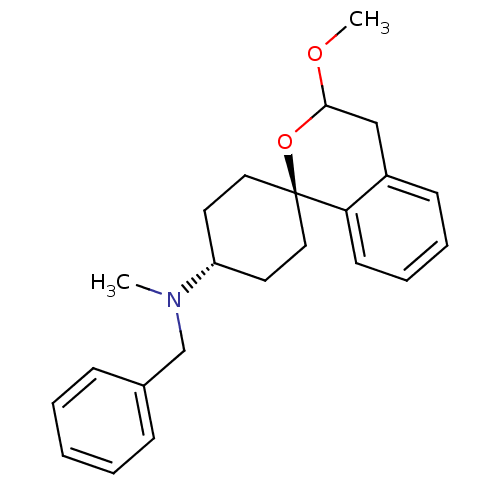
| ||