| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Uracil-DNA glycosylase |
|---|
| Ligand | BDBM50078342 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_212798 (CHEMBL878031) |
|---|
| IC50 | 90000±n/a nM |
|---|
| Citation |  Sun, H; Zhi, C; Wright, GE; Ubiali, D; Pregnolato, M; Verri, A; Focher, F; Spadari, S Molecular modeling and synthesis of inhibitors of herpes simplex virus type 1 uracil-DNA glycosylase. J Med Chem42:2344-50 (1999) [PubMed] Article Sun, H; Zhi, C; Wright, GE; Ubiali, D; Pregnolato, M; Verri, A; Focher, F; Spadari, S Molecular modeling and synthesis of inhibitors of herpes simplex virus type 1 uracil-DNA glycosylase. J Med Chem42:2344-50 (1999) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Uracil-DNA glycosylase |
|---|
| Name: | Uracil-DNA glycosylase |
|---|
| Synonyms: | DGU | UDG | UNG | UNG1 | UNG15 | UNG_HUMAN | Uracil-DNA glycosylase (UNG) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 34661.15 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P1305 |
|---|
| Residue: | 313 |
|---|
| Sequence: | MIGQKTLYSFFSPSPARKRHAPSPEPAVQGTGVAGVPEESGDAAAIPAKKAPAGQEEPGT
PPSSPLSAEQLDRIQRNKAAALLRLAARNVPVGFGESWKKHLSGEFGKPYFIKLMGFVAE
ERKHYTVYPPPHQVFTWTQMCDIKDVKVVILGQDPYHGPNQAHGLCFSVQRPVPPPPSLE
NIYKELSTDIEDFVHPGHGDLSGWAKQGVLLLNAVLTVRAHQANSHKERGWEQFTDAVVS
WLNQNSNGLVFLLWGSYAQKKGSAIDRKRHHVLQTAHPSPLSVYRGFFGCRHFSKTNELL
QKSGKKPIDWKEL
|
|
|
|---|
| BDBM50078342 |
|---|
| n/a |
|---|
| Name | BDBM50078342 |
|---|
| Synonyms: | 6-(4-Hexyl-phenylamino)-1-(3-hydroxy-propyl)-1H-pyrimidine-2,4-dione | CHEMBL80579 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H27N3O3 |
|---|
| Mol. Mass. | 345.436 |
|---|
| SMILES | CCCCCCc1ccc(Nc2cc(=O)[nH]c(=O)n2CCCO)cc1 |
|---|
| Structure | 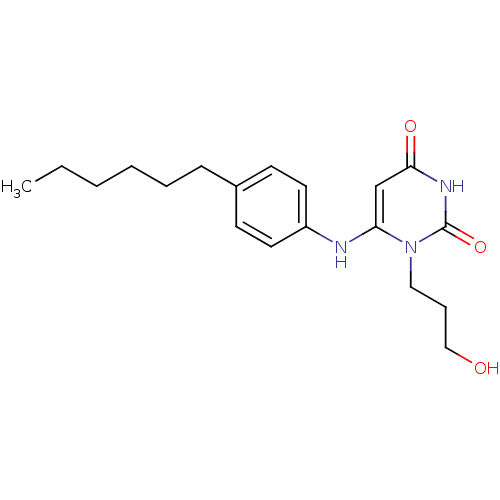
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Sun, H; Zhi, C; Wright, GE; Ubiali, D; Pregnolato, M; Verri, A; Focher, F; Spadari, S Molecular modeling and synthesis of inhibitors of herpes simplex virus type 1 uracil-DNA glycosylase. J Med Chem42:2344-50 (1999) [PubMed] Article
Sun, H; Zhi, C; Wright, GE; Ubiali, D; Pregnolato, M; Verri, A; Focher, F; Spadari, S Molecular modeling and synthesis of inhibitors of herpes simplex virus type 1 uracil-DNA glycosylase. J Med Chem42:2344-50 (1999) [PubMed] Article