| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 60 kDa heat shock protein, mitochondrial |
|---|
| Ligand | BDBM31712 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1891106 (CHEMBL4392933) |
|---|
| IC50 | 1300±n/a nM |
|---|
| Citation |  Stevens, M; Abdeen, S; Salim, N; Ray, AM; Washburn, A; Chitre, S; Sivinski, J; Park, Y; Hoang, QQ; Chapman, E; Johnson, SM HSP60/10 chaperonin systems are inhibited by a variety of approved drugs, natural products, and known bioactive molecules. Bioorg Med Chem Lett29:1106-1112 (2019) [PubMed] Article Stevens, M; Abdeen, S; Salim, N; Ray, AM; Washburn, A; Chitre, S; Sivinski, J; Park, Y; Hoang, QQ; Chapman, E; Johnson, SM HSP60/10 chaperonin systems are inhibited by a variety of approved drugs, natural products, and known bioactive molecules. Bioorg Med Chem Lett29:1106-1112 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 60 kDa heat shock protein, mitochondrial |
|---|
| Name: | 60 kDa heat shock protein, mitochondrial |
|---|
| Synonyms: | 3.6.4.9 | 60 kDa chaperonin | 60 kDa heat shock protein, mitochondrial | CH60_HUMAN | CPN60 | Chaperonin 60 | HSP-60 | HSP60 | HSP60/HSP10 | HSPD1 | Heat shock protein 60 | HuCHA60 | Mitochondrial matrix protein P1 | P60 lymphocyte protein |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 61050.41 |
|---|
| Organism: | Homo sapiens |
|---|
| Description: | ChEMBL_117701 |
|---|
| Residue: | 573 |
|---|
| Sequence: | MLRLPTVFRQMRPVSRVLAPHLTRAYAKDVKFGADARALMLQGVDLLADAVAVTMGPKGR
TVIIEQSWGSPKVTKDGVTVAKSIDLKDKYKNIGAKLVQDVANNTNEEAGDGTTTATVLA
RSIAKEGFEKISKGANPVEIRRGVMLAVDAVIAELKKQSKPVTTPEEIAQVATISANGDK
EIGNIISDAMKKVGRKGVITVKDGKTLNDELEIIEGMKFDRGYISPYFINTSKGQKCEFQ
DAYVLLSEKKISSIQSIVPALEIANAHRKPLVIIAEDVDGEALSTLVLNRLKVGLQVVAV
KAPGFGDNRKNQLKDMAIATGGAVFGEEGLTLNLEDVQPHDLGKVGEVIVTKDDAMLLKG
KGDKAQIEKRIQEIIEQLDVTTSEYEKEKLNERLAKLSDGVAVLKVGGTSDVEVNEKKDR
VTDALNATRAAVEEGIVLGGGCALLRCIPALDSLTPANEDQKIGIEIIKRTLKIPAMTIA
KNAGVEGSLIVEKIMQSSSEVGYDAMAGDFVNMVEKGIIDPTKVVRTALLDAAGVASLLT
TAEVVVTEIPKEEKDPGMGAMGGMGGGMGGGMF
|
|
|
|---|
| BDBM31712 |
|---|
| n/a |
|---|
| Name | BDBM31712 |
|---|
| Synonyms: | HEXACHLOROPHENE | Hexach-lorophene | MLS000028433 | SMR000058356 | cid_3598 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H6Cl6O2 |
|---|
| Mol. Mass. | 406.904 |
|---|
| SMILES | Oc1c(Cl)cc(Cl)c(Cl)c1Cc1c(O)c(Cl)cc(Cl)c1Cl |
|---|
| Structure | 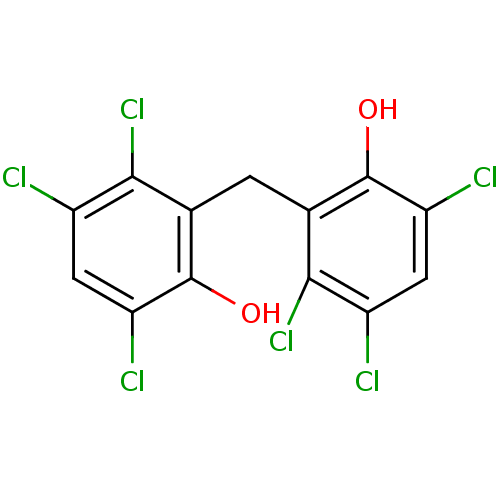
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Stevens, M; Abdeen, S; Salim, N; Ray, AM; Washburn, A; Chitre, S; Sivinski, J; Park, Y; Hoang, QQ; Chapman, E; Johnson, SM HSP60/10 chaperonin systems are inhibited by a variety of approved drugs, natural products, and known bioactive molecules. Bioorg Med Chem Lett29:1106-1112 (2019) [PubMed] Article
Stevens, M; Abdeen, S; Salim, N; Ray, AM; Washburn, A; Chitre, S; Sivinski, J; Park, Y; Hoang, QQ; Chapman, E; Johnson, SM HSP60/10 chaperonin systems are inhibited by a variety of approved drugs, natural products, and known bioactive molecules. Bioorg Med Chem Lett29:1106-1112 (2019) [PubMed] Article