| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Egl nine homolog 1/Prolyl hydroxylase EGLN2/Prolyl hydroxylase EGLN3/Transmembrane prolyl 4-hydroxylase |
|---|
| Ligand | BDBM50529632 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1909275 (CHEMBL4411721) |
|---|
| EC50 | >50000±n/a nM |
|---|
| Citation |  Minassi, A; Rogati, F; Cruz, C; Prados, ME; Galera, N; Jinénez, C; Appendino, G; Bellido, ML; Calzado, MA; Caprioglio, D; Muñoz, E Triterpenoid Hydroxamates as HIF Prolyl Hydrolase Inhibitors. J Nat Prod81:2235-2243 (2018) [PubMed] Article Minassi, A; Rogati, F; Cruz, C; Prados, ME; Galera, N; Jinénez, C; Appendino, G; Bellido, ML; Calzado, MA; Caprioglio, D; Muñoz, E Triterpenoid Hydroxamates as HIF Prolyl Hydrolase Inhibitors. J Nat Prod81:2235-2243 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Egl nine homolog 1/Prolyl hydroxylase EGLN2/Prolyl hydroxylase EGLN3/Transmembrane prolyl 4-hydroxylase |
|---|
| Name: | Egl nine homolog 1/Prolyl hydroxylase EGLN2/Prolyl hydroxylase EGLN3/Transmembrane prolyl 4-hydroxylase |
|---|
| Synonyms: | Hypoxia-inducible factor prolyl hydroxylase |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 1909275 |
|---|
| Components: | This complex has 4 components. |
|---|
| Component 1 |
| Name: | Prolyl hydroxylase EGLN2 |
|---|
| Synonyms: | EGLN2 | EGLN2_HUMAN | EIT6 | Egl nine homolog 2 | Estrogen-induced tag 6 | HIF prolyl hydroxylase | HIF-PH1 | HIF-prolyl hydroxylase 1 | HPH-3 | Hypoxia-inducible factor prolyl hydroxylase | PHD1 | Prolyl hydroxylase domain-containing protein 1 | Prolyl hydroxylase domain-containing protein 1 (PHD1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 43657.01 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q96KS0 |
|---|
| Residue: | 407 |
|---|
| Sequence: | MDSPCQPQPLSQALPQLPGSSSEPLEPEPGRARMGVESYLPCPLLPSYHCPGVPSEASAG
SGTPRATATSTTASPLRDGFGGQDGGELRPLQSEGAAALVTKGCQRLAAQGARPEAPKRK
WAEDGGDAPSPSKRPWARQENQEAEREGGMSCSCSSGSGEASAGLMEEALPSAPERLALD
YIVPCMRYYGICVKDSFLGAALGGRVLAEVEALKRGGRLRDGQLVSQRAIPPRSIRGDQI
AWVEGHEPGCRSIGALMAHVDAVIRHCAGRLGSYVINGRTKAMVACYPGNGLGYVRHVDN
PHGDGRCITCIYYLNQNWDVKVHGGLLQIFPEGRPVVANIEPLFDRLLIFWSDRRNPHEV
KPAYATRYAITVWYFDAKERAAAKDKYQLASGQKGVQVPVSQPPTPT
|
|
|
|---|
| Component 2 |
| Name: | Transmembrane prolyl 4-hydroxylase |
|---|
| Synonyms: | Hypoxia-inducible factor prolyl 4-hydroxylase | Hypoxia-inducible factor prolyl hydroxylase | Hypoxia-inducible factor prolyl hydroxylase 4 | Hypoxia-inducible factor prolyl hydroxylase 4 (HIF) | P4HTM | P4HTM_HUMAN | PH4 | Transmembrane prolyl 4-hydroxylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 56654.25 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9NXG6 |
|---|
| Residue: | 502 |
|---|
| Sequence: | MAAAAVTGQRPETAAAEEASRPQWAPPDHCQAQAAAGLGDGEDAPVRPLCKPRGICSRAY
FLVLMVFVHLYLGNVLALLLFVHYSNGDESSDPGPQHRAQGPGPEPTLGPLTRLEGIKVG
HERKVQLVTDRDHFIRTLSLKPLLFEIPGFLTDEECRLIIHLAQMKGLQRSQILPTEEYE
EAMSTMQVSQLDLFRLLDQNRDGHLQLREVLAQTRLGNGWWMTPESIQEMYAAIKADPDG
DGVLSLQEFSNMDLRDFHKYMRSHKAESSELVRNSHHTWLYQGEGAHHIMRAIRQRVLRL
TRLSPEIVELSEPLQVVRYGEGGHYHAHVDSGPVYPETICSHTKLVANESVPFETSCRYM
TVLFYLNNVTGGGETVFPVADNRTYDEMSLIQDDVDLRDTRRHCDKGNLRVKPQQGTAVF
WYNYLPDGQGWVGDVDDYSLHGGCLVTRGTKWIANNWINVDPSRARQALFQQEMARLARE
GGTDSQPEWALDRAYRDARVEL
|
|
|
|---|
| Component 3 |
| Name: | Egl nine homolog 1 |
|---|
| Synonyms: | C1orf12 | EGLN1 | EGLN1_HUMAN | Egl nine homolog 1 (EGLN1) | Hypoxia-inducible factor prolyl hydroxylase 2 (HIF-PH2) | Hypoxia-inducible factor prolyl hydroxylase 2 (HIFPH2) | Prolyl hydroxylase domain-containing protein 2 (PHD2) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 46035.59 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9GZT9 |
|---|
| Residue: | 426 |
|---|
| Sequence: | MANDSGGPGGPSPSERDRQYCELCGKMENLLRCSRCRSSFYCCKEHQRQDWKKHKLVCQG
SEGALGHGVGPHQHSGPAPPAAVPPPRAGAREPRKAAARRDNASGDAAKGKVKAKPPADP
AAAASPCRAAAGGQGSAVAAEAEPGKEEPPARSSLFQEKANLYPPSNTPGDALSPGGGLR
PNGQTKPLPALKLALEYIVPCMNKHGICVVDDFLGKETGQQIGDEVRALHDTGKFTDGQL
VSQKSDSSKDIRGDKITWIEGKEPGCETIGLLMSSMDDLIRHCNGKLGSYKINGRTKAMV
ACYPGNGTGYVRHVDNPNGDGRCVTCIYYLNKDWDAKVSGGILRIFPEGKAQFADIEPKF
DRLLFFWSDRRNPHEVQPAYATRYAITVWYFDADERARAKVKYLTGEKGVRVELNKPSDS
VGKDVF
|
|
|
|---|
| Component 4 |
| Name: | Prolyl hydroxylase EGLN3 |
|---|
| Synonyms: | EGLN3 | EGLN3_HUMAN | Egl nine homolog 3 (EGLIN3) | Egl nine homolog 3 (EGLN3) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 27265.54 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9H6Z9 |
|---|
| Residue: | 239 |
|---|
| Sequence: | MPLGHIMRLDLEKIALEYIVPCLHEVGFCYLDNFLGEVVGDCVLERVKQLHCTGALRDGQ
LAGPRAGVSKRHLRGDQITWIGGNEEGCEAISFLLSLIDRLVLYCGSRLGKYYVKERSKA
MVACYPGNGTGYVRHVDNPNGDGRCITCIYYLNKNWDAKLHGGILRIFPEGKSFIADVEP
IFDRLLFFWSDRRNPHEVQPSYATRYAMTVWYFDAEERAEAKKKFRNLTRKTESALTED
|
|
|
|---|
| BDBM50529632 |
|---|
| n/a |
|---|
| Name | BDBM50529632 |
|---|
| Synonyms: | CHEMBL4064111 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C30H46O4 |
|---|
| Mol. Mass. | 470.6838 |
|---|
| SMILES | [H][C@@]12CC(C)(C)CC[C@@]1(CC[C@]1(C)C2=CC(=O)[C@]2([H])[C@@]3(C)CC[C@H](O)C(C)(C)[C@]3([H])CC[C@@]12C)C(O)=O |r,t:15| |
|---|
| Structure | 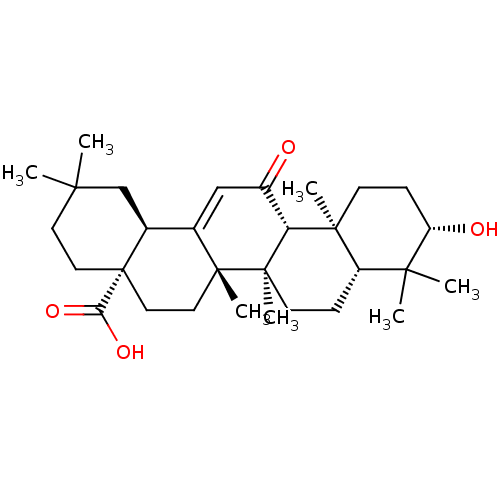
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Minassi, A; Rogati, F; Cruz, C; Prados, ME; Galera, N; Jinénez, C; Appendino, G; Bellido, ML; Calzado, MA; Caprioglio, D; Muñoz, E Triterpenoid Hydroxamates as HIF Prolyl Hydrolase Inhibitors. J Nat Prod81:2235-2243 (2018) [PubMed] Article
Minassi, A; Rogati, F; Cruz, C; Prados, ME; Galera, N; Jinénez, C; Appendino, G; Bellido, ML; Calzado, MA; Caprioglio, D; Muñoz, E Triterpenoid Hydroxamates as HIF Prolyl Hydrolase Inhibitors. J Nat Prod81:2235-2243 (2018) [PubMed] Article