| Reaction Details |
|---|
 | Report a problem with these data |
| Target | AMP deaminase 3 |
|---|
| Ligand | BDBM50087418 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_28867 (CHEMBL644199) |
|---|
| Ki | 2100±n/a nM |
|---|
| Citation |  Bookser, BC; Kasibhatla, SR; Erion, MD AMP deaminase inhibitors. 4. Further N3-substituted coformycin aglycon analogues: N3-alkylmalonates as ribose 5'-monophosphate mimetics. J Med Chem43:1519-24 (2000) [PubMed] Bookser, BC; Kasibhatla, SR; Erion, MD AMP deaminase inhibitors. 4. Further N3-substituted coformycin aglycon analogues: N3-alkylmalonates as ribose 5'-monophosphate mimetics. J Med Chem43:1519-24 (2000) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| AMP deaminase 3 |
|---|
| Name: | AMP deaminase 3 |
|---|
| Synonyms: | AMP deaminase 3 | AMP deaminase 3 (hAMPD3) | AMPD3 | AMPD3_HUMAN |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 88818.80 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 767 |
|---|
| Sequence: | MPRQFPKLNISEVDEQVRLLAEKVFAKVLREEDSKDALSLFTVPEDCPIGQKEAKERELQ
KELAEQKSVETAKRKKSFKMIRSQSLSLQMPPQQDWKGPPAASPAMSPTTPVVTGATSLP
TPAPYAMPEFQRVTISGDYCAGITLEDYEQAAKSLAKALMIREKYARLAYHRFPRITSQY
LGHPRADTAPPEEGLPDFHPPPLPQEDPYCLDDAPPNLDYLVHMQGGILFVYDNKKMLEH
QEPHSLPYPDLETYTVDMSHILALITDGPTKTYCHRRLNFLESKFSLHEMLNEMSEFKEL
KSNPHRDFYNVRKVDTHIHAAACMNQKHLLRFIKHTYQTEPDRTVAEKRGRKITLRQVFD
GLHMDPYDLTVDSLDVHAGRQTFHRFDKFNSKYNPVGASELRDLYLKTENYLGGEYFARM
VKEVARELEESKYQYSEPRLSIYGRSPEEWPNLAYWFIQHKVYSPNMRWIIQVPRIYDIF
RSKKLLPNFGKMLENIFLPLFKATINPQDHRELHLFLKYVTGFDSVDDESKHSDHMFSDK
SPNPDVWTSEQNPPYSYYLYYMYANIMVLNNLRRERGLSTFLFRPHCGEAGSITHLVSAF
LTADNISHGLLLKKSPVLQYLYYLAQIPIAMSPLSNNSLFLEYSKNPLREFLHKGLHVSL
STDDPMQFHYTKEALMEEYAIAAQVWKLSTCDLCEIARNSVLQSGLSHQEKQKFLGQNYY
KEGPEGNDIRKTNVAQIRMAFRYETLCNELSFLSDAMKSEEITALTN
|
|
|
|---|
| BDBM50087418 |
|---|
| n/a |
|---|
| Name | BDBM50087418 |
|---|
| Synonyms: | 6-(8-Hydroxy-7,8-dihydro-6H-imidazo[4,5-d][1,3]diazepin-3-yl)-2-methyl-2-phenethylcarbamoyl-hexanoic acid 0.75H2O.0.3CH3CO2H | CHEMBL287822 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H29N5O4 |
|---|
| Mol. Mass. | 427.4968 |
|---|
| SMILES | CC(CCCCn1cnc2C(O)CNC=Nc12)(C(O)=O)C(=O)NCCc1ccccc1 |c:14| |
|---|
| Structure | 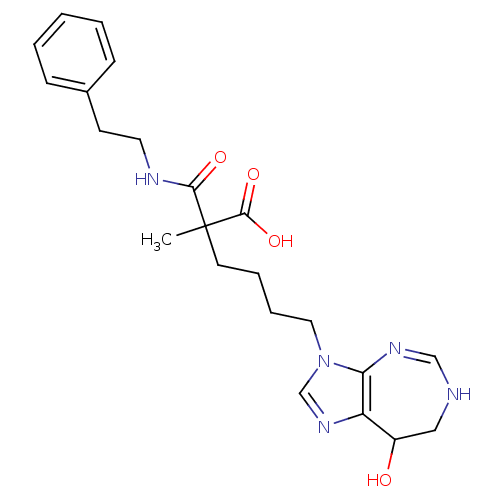
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bookser, BC; Kasibhatla, SR; Erion, MD AMP deaminase inhibitors. 4. Further N3-substituted coformycin aglycon analogues: N3-alkylmalonates as ribose 5'-monophosphate mimetics. J Med Chem43:1519-24 (2000) [PubMed]
Bookser, BC; Kasibhatla, SR; Erion, MD AMP deaminase inhibitors. 4. Further N3-substituted coformycin aglycon analogues: N3-alkylmalonates as ribose 5'-monophosphate mimetics. J Med Chem43:1519-24 (2000) [PubMed]