

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Muscarinic acetylcholine receptor M3 | ||
| Ligand | BDBM50538207 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1972409 (CHEMBL4605227) | ||
| Ki | 0.350000±n/a nM | ||
| Citation |  Fischer, O; Hofmann, J; Rampp, H; Kaindl, J; Pratsch, G; Bartuschat, A; Taudte, RV; Fromm, MF; Hübner, H; Gmeiner, P; Heinrich, MR Regiospecific Introduction of Halogens on the 2-Aminobiphenyl Subunit Leading to Highly Potent and Selective M3 Muscarinic Acetylcholine Receptor Antagonists and Weak Inverse Agonists. J Med Chem63:4349-4369 (2020) [PubMed] Article Fischer, O; Hofmann, J; Rampp, H; Kaindl, J; Pratsch, G; Bartuschat, A; Taudte, RV; Fromm, MF; Hübner, H; Gmeiner, P; Heinrich, MR Regiospecific Introduction of Halogens on the 2-Aminobiphenyl Subunit Leading to Highly Potent and Selective M3 Muscarinic Acetylcholine Receptor Antagonists and Weak Inverse Agonists. J Med Chem63:4349-4369 (2020) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Muscarinic acetylcholine receptor M3 | |||
| Name: | Muscarinic acetylcholine receptor M3 | ||
| Synonyms: | ACM3_HUMAN | CHRM3 | Cholinergic, muscarinic M3 | Muscarinic Receptors M3 | Muscarinic receptor M3 | RecName: Full=Muscarinic acetylcholine receptor M3 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 66151.03 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P20309 | ||
| Residue: | 590 | ||
| Sequence: |
| ||
| BDBM50538207 | |||
| n/a | |||
| Name | BDBM50538207 | ||
| Synonyms: | CHEMBL4640324 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H19ClF2N2O2 | ||
| Mol. Mass. | 392.827 | ||
| SMILES | Fc1ccc(NC(=O)O[C@H]2CN3CCC2CC3)c(c1)-c1ccc(F)c(Cl)c1 |r,wU:9.8,(12.23,-40.25,;12.23,-38.74,;13.58,-37.97,;13.57,-36.42,;12.23,-35.65,;12.23,-34.11,;13.56,-33.34,;13.56,-31.8,;14.89,-34.11,;16.23,-33.33,;16.23,-31.8,;17.56,-31.03,;18.9,-31.8,;18.9,-33.34,;17.57,-34.11,;18.04,-32.92,;17.05,-32.52,;10.91,-36.42,;10.9,-37.97,;9.57,-35.66,;8.25,-36.42,;6.91,-35.66,;6.91,-34.11,;5.6,-33.36,;8.25,-33.34,;8.25,-31.83,;9.57,-34.11,)| | ||
| Structure | 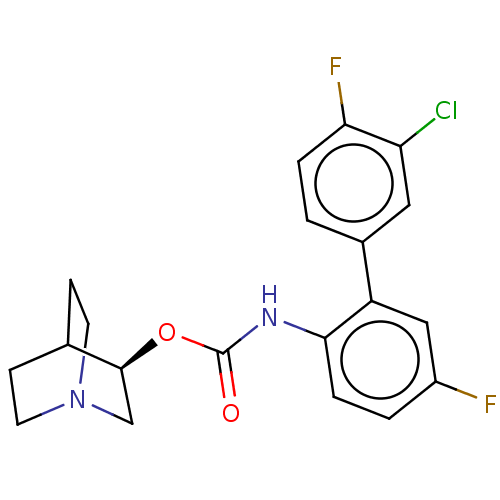
| ||