| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 8 |
|---|
| Ligand | BDBM50538663 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1973335 (CHEMBL4606153) |
|---|
| IC50 | 44±n/a nM |
|---|
| Citation |  Tng, J; Lim, J; Wu, KC; Lucke, AJ; Xu, W; Reid, RC; Fairlie, DP Achiral Derivatives of Hydroxamate AR-42 Potently Inhibit Class I HDAC Enzymes and Cancer Cell Proliferation. J Med Chem63:5956-5971 (2020) [PubMed] Article Tng, J; Lim, J; Wu, KC; Lucke, AJ; Xu, W; Reid, RC; Fairlie, DP Achiral Derivatives of Hydroxamate AR-42 Potently Inhibit Class I HDAC Enzymes and Cancer Cell Proliferation. J Med Chem63:5956-5971 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 8 |
|---|
| Name: | Histone deacetylase 8 |
|---|
| Synonyms: | HD8 | HDAC8 | HDAC8_HUMAN | HDACL1 | Histone deacetylase 8 (HDAC-8) | Human HDAC8 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 41749.60 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9BY41 |
|---|
| Residue: | 377 |
|---|
| Sequence: | MEEPEEPADSGQSLVPVYIYSPEYVSMCDSLAKIPKRASMVHSLIEAYALHKQMRIVKPK
VASMEEMATFHTDAYLQHLQKVSQEGDDDHPDSIEYGLGYDCPATEGIFDYAAAIGGATI
TAAQCLIDGMCKVAINWSGGWHHAKKDEASGFCYLNDAVLGILRLRRKFERILYVDLDLH
HGDGVEDAFSFTSKVMTVSLHKFSPGFFPGTGDVSDVGLGKGRYYSVNVPIQDGIQDEKY
YQICESVLKEVYQAFNPKAVVLQLGADTIAGDPMCSFNMTPVGIGKCLKYILQWQLATLI
LGGGGYNLANTARCWTYLTGVILGKTLSSEIPDHEFFTAYGPDYVLEITPSCRPDRNEPH
RIQQILNYIKGNLKHVV
|
|
|
|---|
| BDBM50538663 |
|---|
| n/a |
|---|
| Name | BDBM50538663 |
|---|
| Synonyms: | CHEMBL4642874 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H22N2O4 |
|---|
| Mol. Mass. | 390.4318 |
|---|
| SMILES | ONC(=O)c1ccc(NC(=O)C2(CCOCC2)c2ccc3ccccc3c2)cc1 |
|---|
| Structure | 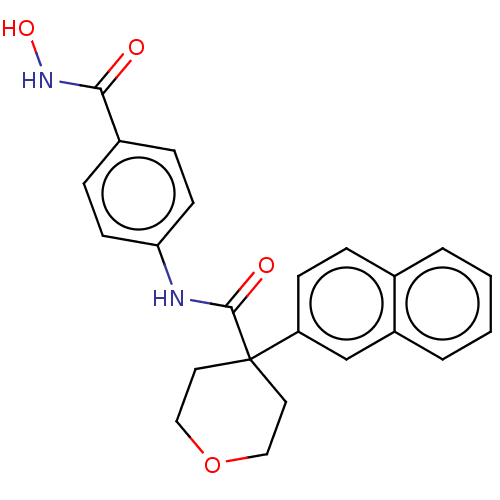
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Tng, J; Lim, J; Wu, KC; Lucke, AJ; Xu, W; Reid, RC; Fairlie, DP Achiral Derivatives of Hydroxamate AR-42 Potently Inhibit Class I HDAC Enzymes and Cancer Cell Proliferation. J Med Chem63:5956-5971 (2020) [PubMed] Article
Tng, J; Lim, J; Wu, KC; Lucke, AJ; Xu, W; Reid, RC; Fairlie, DP Achiral Derivatives of Hydroxamate AR-42 Potently Inhibit Class I HDAC Enzymes and Cancer Cell Proliferation. J Med Chem63:5956-5971 (2020) [PubMed] Article