

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Poly [ADP-ribose] polymerase 2 | ||
| Ligand | BDBM50541697 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1985251 (CHEMBL4618657) | ||
| IC50 | >10000±n/a nM | ||
| Citation |  Waaler, J; Leenders, RGG; Sowa, ST; Alam Brinch, S; Lycke, M; Nieczypor, P; Aertssen, S; Murthy, S; Galera-Prat, A; Damen, E; Wegert, A; Nazaré, M; Lehtiö, L; Krauss, S Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor. J Med Chem63:6834-6846 (2020) [PubMed] Article Waaler, J; Leenders, RGG; Sowa, ST; Alam Brinch, S; Lycke, M; Nieczypor, P; Aertssen, S; Murthy, S; Galera-Prat, A; Damen, E; Wegert, A; Nazaré, M; Lehtiö, L; Krauss, S Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor. J Med Chem63:6834-6846 (2020) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Poly [ADP-ribose] polymerase 2 | |||
| Name: | Poly [ADP-ribose] polymerase 2 | ||
| Synonyms: | (ARTD2 or PARP2) | ADPRT2 | ADPRTL2 | PARP2 | PARP2_HUMAN | Poly [ADP-ribose] polymerase 2 (PARP-2) | Poly [ADP-ribose] polymerase 2 (PARP2) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 66225.70 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q9UGN5 | ||
| Residue: | 583 | ||
| Sequence: |
| ||
| BDBM50541697 | |||
| n/a | |||
| Name | BDBM50541697 | ||
| Synonyms: | CHEMBL4633637 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H23FN6O2 | ||
| Mol. Mass. | 458.4875 | ||
| SMILES | CCOc1ccc(nc1)-c1nnc([C@H]2C[C@@H](C2)NC(=O)c2ccccn2)n1-c1ccccc1F |r,wU:15.18,wD:13.13,(2.31,-18.95,;3.77,-18.47,;4.92,-19.5,;6.38,-19.02,;7.54,-20.05,;9,-19.57,;9.31,-18.07,;8.17,-17.04,;6.7,-17.51,;10.77,-17.6,;11.25,-16.13,;12.79,-16.13,;13.26,-17.6,;14.73,-18.07,;15.42,-19.45,;16.8,-18.75,;16.1,-17.38,;18.26,-19.23,;19.41,-18.2,;19.09,-16.69,;20.87,-18.67,;22,-17.64,;23.47,-18.11,;23.79,-19.62,;22.65,-20.65,;21.19,-20.18,;12.02,-18.5,;12.02,-20.04,;10.68,-20.81,;10.68,-22.34,;12.02,-23.12,;13.36,-22.34,;13.35,-20.8,;14.68,-20.03,)| | ||
| Structure | 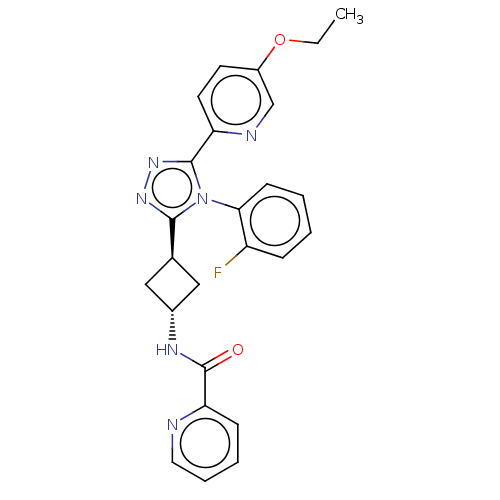
| ||