| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Endoplasmic reticulum aminopeptidase 2 |
|---|
| Ligand | BDBM50331042 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1992033 (CHEMBL4625768) |
|---|
| IC50 | 1259±n/a nM |
|---|
| Citation |  Mpakali, A; Saridakis, E; Giastas, P; Maben, Z; Stern, LJ; Larhed, M; Hallberg, M; Stratikos, E Structural Basis of Inhibition of Insulin-Regulated Aminopeptidase by a Macrocyclic Peptidic Inhibitor. ACS Med Chem Lett11:1429-1434 (2020) [PubMed] Article Mpakali, A; Saridakis, E; Giastas, P; Maben, Z; Stern, LJ; Larhed, M; Hallberg, M; Stratikos, E Structural Basis of Inhibition of Insulin-Regulated Aminopeptidase by a Macrocyclic Peptidic Inhibitor. ACS Med Chem Lett11:1429-1434 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Endoplasmic reticulum aminopeptidase 2 |
|---|
| Name: | Endoplasmic reticulum aminopeptidase 2 |
|---|
| Synonyms: | Aminopeptidase | ERAP2 | ERAP2_HUMAN | LRAP |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 110463.90 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1473169 |
|---|
| Residue: | 960 |
|---|
| Sequence: | MFHSSAMVNSHRKPMFNIHRGFYCLTAILPQICICSQFSVPSSYHFTEDPGAFPVATNGE
RFPWQELRLPSVVIPLHYDLFVHPNLTSLDFVASEKIEVLVSNATQFIILHSKDLEITNA
TLQSEEDSRYMKPGKELKVLSYPAHEQIALLVPEKLTPHLKYYVAMDFQAKLGDGFEGFY
KSTYRTLGGETRILAVTDFEPTQARMAFPCFDEPLFKANFSIKIRRESRHIALSNMPKVK
TIELEGGLLEDHFETTVKMSTYLVAYIVCDFHSLSGFTSSGVKVSIYASPDKRNQTHYAL
QASLKLLDFYEKYFDIYYPLSKLDLIAIPDFAPGAMENWGLITYRETSLLFDPKTSSASD
KLWVTRVIAHELAHQWFGNLVTMEWWNDIWLKEGFAKYMELIAVNATYPELQFDDYFLNV
CFEVITKDSLNSSRPISKPAETPTQIQEMFDEVSYNKGACILNMLKDFLGEEKFQKGIIQ
YLKKFSYRNAKNDDLWSSLSNSCLESDFTSGGVCHSDPKMTSNMLAFLGENAEVKEMMTT
WTLQKGIPLLVVKQDGCSLRLQQERFLQGVFQEDPEWRALQERYLWHIPLTYSTSSSNVI
HRHILKSKTDTLDLPEKTSWVKFNVDSNGYYIVHYEGHGWDQLITQLNQNHTLLRPKDRV
GLIHDVFQLVGAGRLTLDKALDMTYYLQHETSSPALLEGLSYLESFYHMMDRRNISDISE
NLKRYLLQYFKPVIDRQSWSDKGSVWDRMLRSALLKLACDLNHAPCIQKAAELFSQWMES
SGKLNIPTDVLKIVYSVGAQTTAGWNYLLEQYELSMSSAEQNKILYALSTSKHQEKLLKL
IELGMEGKVIKTQNLAALLHAIARRPKGQQLAWDFVRENWTHLLKKFDLGSYDIRMIISG
TTAHFSSKDKLQEVKLFFESLEAQGSHLDIFQTVLETITKNIKWLEKNLPTLRTWLMVNT
|
|
|
|---|
| BDBM50331042 |
|---|
| n/a |
|---|
| Name | BDBM50331042 |
|---|
| Synonyms: | 2-(2-(((4R,8S,11S)-11-amino-8-(4-hydroxybenzyl)-6,10-dioxo-1,2-dithia-5,9-diazacyclotridecane-4-carboxamido)methyl)phenyl)acetic acid | CHEMBL1277623 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H32N4O6S2 |
|---|
| Mol. Mass. | 560.685 |
|---|
| SMILES | N[C@H]1CCSSC[C@H](NC(=O)C[C@H](Cc2ccc(O)cc2)NC1=O)C(=O)NCc1ccccc1CC(O)=O |r| |
|---|
| Structure | 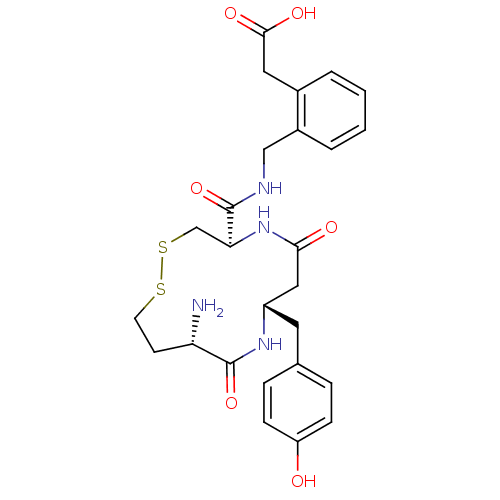
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Mpakali, A; Saridakis, E; Giastas, P; Maben, Z; Stern, LJ; Larhed, M; Hallberg, M; Stratikos, E Structural Basis of Inhibition of Insulin-Regulated Aminopeptidase by a Macrocyclic Peptidic Inhibitor. ACS Med Chem Lett11:1429-1434 (2020) [PubMed] Article
Mpakali, A; Saridakis, E; Giastas, P; Maben, Z; Stern, LJ; Larhed, M; Hallberg, M; Stratikos, E Structural Basis of Inhibition of Insulin-Regulated Aminopeptidase by a Macrocyclic Peptidic Inhibitor. ACS Med Chem Lett11:1429-1434 (2020) [PubMed] Article