| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Dual specificity tyrosine-phosphorylation-regulated kinase 1A |
|---|
| Ligand | BDBM50249773 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2012564 (CHEMBL4666142) |
|---|
| IC50 | 20±n/a nM |
|---|
| Citation |  Tazarki, H; Zeinyeh, W; Esvan, YJ; Knapp, S; Chatterjee, D; Schröder, M; Joerger, AC; Khiari, J; Josselin, B; Baratte, B; Bach, S; Ruchaud, S; Anizon, F; Giraud, F; Moreau, P New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis. Eur J Med Chem166:304-317 (2019) [PubMed] Article Tazarki, H; Zeinyeh, W; Esvan, YJ; Knapp, S; Chatterjee, D; Schröder, M; Joerger, AC; Khiari, J; Josselin, B; Baratte, B; Bach, S; Ruchaud, S; Anizon, F; Giraud, F; Moreau, P New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis. Eur J Med Chem166:304-317 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Dual specificity tyrosine-phosphorylation-regulated kinase 1A |
|---|
| Name: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A |
|---|
| Synonyms: | DYR1A_RAT | Dual specificity tyrosine-phosphorylation-regulated kinase 1A | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | Dyrk | Dyrk1a |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 85573.59 |
|---|
| Organism: | RAT |
|---|
| Description: | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A 0 RAT::Q63470 |
|---|
| Residue: | 763 |
|---|
| Sequence: | MHTGGETSACKPSSVRLAPSFSFHAAGLQMAAQMPHSHQYSDRRQPNISDQQVSALSYSD
QIQQPLTNQVMPDIVMLQRRMPQTFRDPATAPLRKLSVDLIKTYKHINEVYYAKKKRRHQ
QGQGDDSSHKKERKVYNDGYDDDNYDYIVKNGEKWMDRYEIDSLIGKGSFGQVVKAYDRV
EQEWVAIKIIKNKKAFLNQAQIEVRLLELMNKHDTEMKYYIVHLKRHFMFRNHLCLVFEM
LSYNLYDLLRNTNFRGVSLNLTRKFAQQMCTALLFLATPELSIIHCDLKPENILLCNPKR
SAIKIVDFGSSCQLGQRIYQYIQSRFYRSPEVLLGMPYDLAIDMWSLGCILVEMHTGEPL
FSGANEVDQMNKIVEVLGIPPAHILDQAPKARKFFEKLPDGTWSLKKTKDGKREYKPPGT
RKLHNILGVETGGPGGRRAGESGHTVADYLKFKDLILRMLDYDPKTRIQPYYALQHSFFK
KTADEGTNTSNSVSTSPAMEQSQSSGTTSSTSSSSGGSSGTSNSGRARSDPTHQHRHSGG
HFAAAVQAMDCETHSPQVRQQFPAPLGWSGTEAPTQVTVETHPVQETTFHVAPQQNALHH
HHGNSSHHHHHHHHHHHHHGQQALGNRTRPRVYNSPTNSSSTQDSMEVGHSHHSMTSLSS
STTSSSTSSSSTGNQGNQAYQNRPVAANTLDFGQNGAMDVNLTVYSNPRQETGIAGHPTY
QFSANTGPAHYMTEGHLTMRQGADREESPMTGVCVQQSPVASS
|
|
|
|---|
| BDBM50249773 |
|---|
| n/a |
|---|
| Name | BDBM50249773 |
|---|
| Synonyms: | CHEMBL4763099 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H25N7 |
|---|
| Mol. Mass. | 351.4487 |
|---|
| SMILES | CN1CCN(CCCNc2ncc3cc4cnccc4c(N)c3n2)CC1 |
|---|
| Structure | 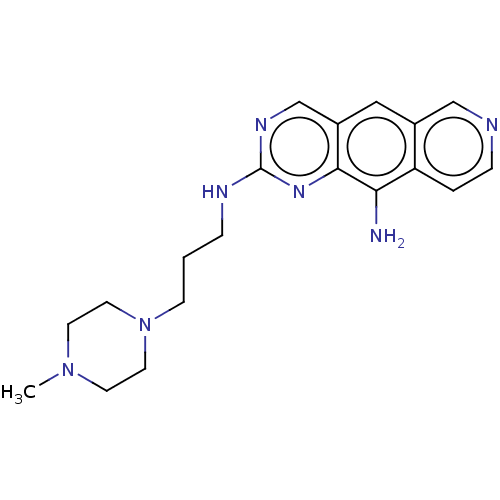
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Tazarki, H; Zeinyeh, W; Esvan, YJ; Knapp, S; Chatterjee, D; Schröder, M; Joerger, AC; Khiari, J; Josselin, B; Baratte, B; Bach, S; Ruchaud, S; Anizon, F; Giraud, F; Moreau, P New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis. Eur J Med Chem166:304-317 (2019) [PubMed] Article
Tazarki, H; Zeinyeh, W; Esvan, YJ; Knapp, S; Chatterjee, D; Schröder, M; Joerger, AC; Khiari, J; Josselin, B; Baratte, B; Bach, S; Ruchaud, S; Anizon, F; Giraud, F; Moreau, P New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis. Eur J Med Chem166:304-317 (2019) [PubMed] Article