| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA gyrase subunit A/B |
|---|
| Ligand | BDBM50249796 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2012762 (CHEMBL4666340) |
|---|
| IC50 | 99±n/a nM |
|---|
| Citation |  Tiz, DB; Skok, ?; Durcik, M; Toma?i?, T; Ma?i?, LP; Ila?, J; Zega, A; Draskovits, G; Révész, T; Nyerges, Á; Pál, C; Cruz, CD; Tammela, P; ?igon, D; Kikelj, D; Zidar, N An optimised series of substituted N-phenylpyrrolamides as DNA gyrase B inhibitors. Eur J Med Chem167:269-290 (2019) [PubMed] Article Tiz, DB; Skok, ?; Durcik, M; Toma?i?, T; Ma?i?, LP; Ila?, J; Zega, A; Draskovits, G; Révész, T; Nyerges, Á; Pál, C; Cruz, CD; Tammela, P; ?igon, D; Kikelj, D; Zidar, N An optimised series of substituted N-phenylpyrrolamides as DNA gyrase B inhibitors. Eur J Med Chem167:269-290 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| DNA gyrase subunit A/B |
|---|
| Name: | DNA gyrase subunit A/B |
|---|
| Synonyms: | DNA Gyrase |
|---|
| Type: | A2B2 tetramer |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | n/a |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | DNA gyrase subunit A |
|---|
| Synonyms: | DNA gyrase subunit A (gyrA) | GYRA_STAAU | gyrA |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 99588.82 |
|---|
| Organism: | Staphylococcus aureus |
|---|
| Description: | n/a |
|---|
| Residue: | 889 |
|---|
| Sequence: | MAELPQSRINERNITSEMRESFLDYAMSVIVARALPDVRDGLKPVHRRILYGLNEQGMTP
DKSYKKSARIVGDVMGKYHPHGDSSIYEAMVRMAQDFSYRYPLVDGQGNFGSMDGDGAAA
MRYTEARMTKITLELLRDINKDTIDFIDNYDGNEREPSVLPARFPNLLANGASGIAVGMA
TNIPPHNLTELINGVLSLSKNPDISIAELMEDIEGPDFPTAGLILGKSGIRRAYETGRGS
IQMRSRAVIEERGGGRQRIVVTEIPFQVNKARMIEKIAELVRDKKIDGITDLRDETSLRT
GVRVVIDVRKDANASVILNNLYKQTPLQTSFGVNMIALVNGRPKLINLKEALVHYLEHQK
TVVRRRTQYNLRKAKDRAHILEGLRIALDHIDEIISTIRESDTDKVAMESLQQRFKLSEK
QAQAILDMRLRRLTGLERDKIEAEYNELLNYISELEAILADEEVLLQLVRDELTEIRDRF
GDDRRTEIQLGGFEDLEDEDLIPEEQIVITLSHNNYIKRLPVSTYRAQNRGGRGVQGMNT
LEEDFVSQLVTLSTHDHVLFFTNKGRVYKLKGYEVPELSRQSKGIPVVNAIELENDEVIS
TMIAVKDLESEDNFLVFATKRGVVKRSALSNFSRINRNGKIAISFREDDELIAVRLTSGQ
EDILIGTSHASLIRFPESTLRPLGRTATGVKGITLREGDEVVGLDVAHANSVDEVLVVTE
NGYGKRTPVNDYRLSNRGGKGIKTATITERNGNVVCITTVTGEEDLMIVTNAGVIIRLDV
ADISQNGRAAQGVRLIRLGDDQFVSTVAKVKEDAEDETNEDEQSTSTVSEDGTEQQREAV
VNDETPGNAIHTEVIDSEENDEDGRIEVRQDFMDRVEEDIQQSLDEDEE
|
|
|
|---|
| Component 2 |
| Name: | DNA gyrase subunit B |
|---|
| Synonyms: | DNA gyrase | DNA gyrase subunit B (DNA gyraseB) | DNA gyrase subunit B (gyrB) | GYRB_STAAU | gyrB |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 72530.91 |
|---|
| Organism: | Staphylococcus aureus |
|---|
| Description: | n/a |
|---|
| Residue: | 644 |
|---|
| Sequence: | MVTALSDVNNTDNYGAGQIQVLEGLEAVRKRPGMYIGSTSERGLHHLVWEIVDNSIDEAL
AGYANQIEVVIEKDNWIKVTDNGRGIPVDIQEKMGRPAVEVILTVLHAGGKFGGGGYKVS
GGLHGVGSSVVNALSQDLEVYVHRNETIYHQAYKKGVPQFDLKEVGTTDKTGTVIRFKAD
GEIFTETTVYNYETLQQRIRELAFLNKGIQITLRDERDEENVREDSYHYEGGIKSYVELL
NENKEPIHDEPIYIHQSKDDIEVEIAIQYNSGYATNLLTYANNIHTYEGGTHEDGFKRAL
TRVLNSYGLSSKIMKEEKDRLSGEDTREGMTAIISIKHGDPQFEGQTKTKLGNSEVRQVV
DKLFSEHFERFLYENPQVARTVVEKGIMAARARVAAKKAREVTRRKSALDVASLPGKLAD
CSSKSPEECEIFLVEGDSAGGSTKSGRDSRTQAILPLRGKILNVEKARLDRILNNNEIRQ
MITAFGTGIGGDFDLAKARYHKIVIMTDADVDGAHIRTLLLTFFYRFMRPLIEAGYVYIA
QPPLYKLTQGKQKYYVYNDRELDKLKSELNPTPKWSIARYKGLGEMNADQLWETTMNPEH
RALLQVKLEDAIEADQTFEMLMGDVVENRRQFIEDNAVYANLDF
|
|
|
|---|
| BDBM50249796 |
|---|
| n/a |
|---|
| Name | BDBM50249796 |
|---|
| Synonyms: | CHEMBL4752072 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H25Cl3N4O5 |
|---|
| Mol. Mass. | 519.806 |
|---|
| SMILES | Cl.C[C@H](NC(=O)c1ccc(NC(=O)c2[nH]c(C)c(Cl)c2Cl)c(OC2CCNCC2)c1)C(O)=O |r| |
|---|
| Structure | 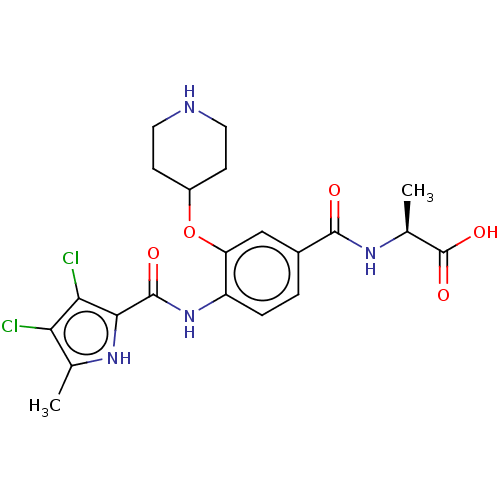
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Tiz, DB; Skok, ?; Durcik, M; Toma?i?, T; Ma?i?, LP; Ila?, J; Zega, A; Draskovits, G; Révész, T; Nyerges, Á; Pál, C; Cruz, CD; Tammela, P; ?igon, D; Kikelj, D; Zidar, N An optimised series of substituted N-phenylpyrrolamides as DNA gyrase B inhibitors. Eur J Med Chem167:269-290 (2019) [PubMed] Article
Tiz, DB; Skok, ?; Durcik, M; Toma?i?, T; Ma?i?, LP; Ila?, J; Zega, A; Draskovits, G; Révész, T; Nyerges, Á; Pál, C; Cruz, CD; Tammela, P; ?igon, D; Kikelj, D; Zidar, N An optimised series of substituted N-phenylpyrrolamides as DNA gyrase B inhibitors. Eur J Med Chem167:269-290 (2019) [PubMed] Article