| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Insulin-like growth factor 1 receptor |
|---|
| Ligand | BDBM50334594 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2025671 (CHEMBL4679484) |
|---|
| IC50 | 160±n/a nM |
|---|
| Citation |  Groendyke, BJ; Nabet, B; Mohardt, ML; Zhang, H; Peng, K; Koide, E; Coffey, CR; Che, J; Scott, DA; Bass, AJ; Gray, NS Discovery of a Pyrimidothiazolodiazepinone as a Potent and Selective Focal Adhesion Kinase (FAK) Inhibitor. ACS Med Chem Lett12:30-38 (2021) [PubMed] Article Groendyke, BJ; Nabet, B; Mohardt, ML; Zhang, H; Peng, K; Koide, E; Coffey, CR; Che, J; Scott, DA; Bass, AJ; Gray, NS Discovery of a Pyrimidothiazolodiazepinone as a Potent and Selective Focal Adhesion Kinase (FAK) Inhibitor. ACS Med Chem Lett12:30-38 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Insulin-like growth factor 1 receptor |
|---|
| Name: | Insulin-like growth factor 1 receptor |
|---|
| Synonyms: | CD_antigen=CD221 | IGF-I receptor | IGF1R | IGF1R_HUMAN | Insulin-like growth factor 1 receptor (IGF1R) | Insulin-like growth factor 1 receptor (IGFIR) | Insulin-like growth factor 1 receptor alpha chain | Insulin-like growth factor 1 receptor beta chain | Insulin-like growth factor I receptor | Insulin-like growth factor receptor (IGFR) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 154776.79 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P08069 |
|---|
| Residue: | 1367 |
|---|
| Sequence: | MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLH
ILLISKAEDYRSYRFPKLTVITEYLLLFRVAGLESLGDLFPNLTVIRGWKLFYNYALVIF
EMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD
LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKMCPSTCGKRACTENNECCHPECLGSCS
APDNDTACVACRHYYYAGVCVPACPPNTYRFEGWRCVDRDFCANILSAESSDSEGFVIHD
GECMQECPSGFIRNGSQSMYCIPCEGPCPKVCEEEKKTKTIDSVTSAQMLQGCTIFKGNL
LINIRRGNNIASELENFMGLIEVVTGYVKIRHSHALVSLSFLKNLRLILGEEQLEGNYSF
YVLDNQNLQQLWDWDHRNLTIKAGKMYFAFNPKLCVSEIYRMEEVTGTKGRQSKGDINTR
NNGERASCESDVLHFTSTTTSKNRIIITWHRYRPPDYRDLISFTVYYKEAPFKNVTEYDG
QDACGSNSWNMVDVDLPPNKDVEPGILLHGLKPWTQYAVYVKAVTLTMVENDHIRGAKSE
ILYIRTNASVPSIPLDVLSASNSSSQLIVKWNPPSLPNGNLSYYIVRWQRQPQDGYLYRH
NYCSKDKIPIRKYADGTIDIEEVTENPKTEVCGGEKGPCCACPKTEAEKQAEKEEAEYRK
VFENFLHNSIFVPRPERKRRDVMQVANTTMSSRSRNTTAADTYNITDPEELETEYPFFES
RVDNKERTVISNLRPFTLYRIDIHSCNHEAEKLGCSASNFVFARTMPAEGADDIPGPVTW
EPRPENSIFLKWPEPENPNGLILMYEIKYGSQVEDQRECVSRQEYRKYGGAKLNRLNPGN
YTARIQATSLSGNGSWTDPVFFYVQAKTGYENFIHLIIALPVAVLLIVGGLVIMLYVFHR
KRNNSRLGNGVLYASVNPEYFSAADVYVPDEWEVAREKITMSRELGQGSFGMVYEGVAKG
VVKDEPETRVAIKTVNEAASMRERIEFLNEASVMKEFNCHHVVRLLGVVSQGQPTLVIME
LMTRGDLKSYLRSLRPEMENNPVLAPPSLSKMIQMAGEIADGMAYLNANKFVHRDLAARN
CMVAEDFTVKIGDFGMTRDIYETDYYRKGGKGLLPVRWMSPESLKDGVFTTYSDVWSFGV
VLWEIATLAEQPYQGLSNEQVLRFVMEGGLLDKPDNCPDMLFELMRMCWQYNPKMRPSFL
EIISSIKEEMEPGFREVSFYYSEENKLPEPEELDLEPENMESVPLDPSASSSSLPLPDRH
SGHKAENGPGPGVLVLRASFDERQPYAHMNGGRKNERALPLPQSSTC
|
|
|
|---|
| BDBM50334594 |
|---|
| n/a |
|---|
| Name | BDBM50334594 |
|---|
| Synonyms: | 2-(5-chloro-2-(2-methoxy-4-morpholinophenylamino)pyrimidin-4-ylamino)-N-methylbenzamide | 2-({5-CHLORO-2-[(2-METHOXY-4-MORPHOLIN-4-YLPHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)-N-METHYLBENZAMIDE | 2-[5-chloro-2-[2-methoxy-4-(4-morpholinyl)phenylamino]pyrimidin-4-ylamino]-N-methylbenzamide | CHEMBL458997 | TAE226 | US10266549, Example 12 | US10774092, Example 12 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H25ClN6O3 |
|---|
| Mol. Mass. | 468.936 |
|---|
| SMILES | CNC(=O)c1ccccc1Nc1nc(Nc2ccc(cc2OC)N2CCOCC2)ncc1Cl |
|---|
| Structure | 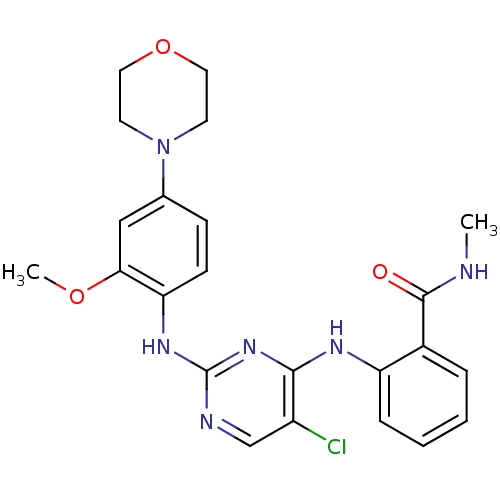
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Groendyke, BJ; Nabet, B; Mohardt, ML; Zhang, H; Peng, K; Koide, E; Coffey, CR; Che, J; Scott, DA; Bass, AJ; Gray, NS Discovery of a Pyrimidothiazolodiazepinone as a Potent and Selective Focal Adhesion Kinase (FAK) Inhibitor. ACS Med Chem Lett12:30-38 (2021) [PubMed] Article
Groendyke, BJ; Nabet, B; Mohardt, ML; Zhang, H; Peng, K; Koide, E; Coffey, CR; Che, J; Scott, DA; Bass, AJ; Gray, NS Discovery of a Pyrimidothiazolodiazepinone as a Potent and Selective Focal Adhesion Kinase (FAK) Inhibitor. ACS Med Chem Lett12:30-38 (2021) [PubMed] Article