| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prostaglandin G/H synthase 2 |
|---|
| Ligand | BDBM50112547 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_159920 |
|---|
| IC50 | 47±n/a nM |
|---|
| Citation |  Kozak, KR; Prusakiewicz, JJ; Rowlinson, SW; Marnett, LJ Enantiospecific, selective cyclooxygenase-2 inhibitors. Bioorg Med Chem Lett12:1315-8 (2002) [PubMed] Kozak, KR; Prusakiewicz, JJ; Rowlinson, SW; Marnett, LJ Enantiospecific, selective cyclooxygenase-2 inhibitors. Bioorg Med Chem Lett12:1315-8 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prostaglandin G/H synthase 2 |
|---|
| Name: | Prostaglandin G/H synthase 2 |
|---|
| Synonyms: | COX2 | Cyclooxygenase | Cyclooxygenase 2 (COX-2) | Cyclooxygenase-2 | Cyclooxygenase-2 (COX-2 AA) | Cyclooxygenase-2 (COX-2 AEA) | Cyclooxygenase-2 (COX-2) | PGH synthase 2 | PGH2_HUMAN | PGHS-2 | PHS II | PTGS2 | Prostaglandin E synthase/G/H synthase 2 | Prostaglandin H2 synthase 2 | Prostaglandin-endoperoxide synthase 2 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 69003.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Recombinant Cox-2 provided by Cayman (Cayman Chemical Co.,Ann Arbor, MI). |
|---|
| Residue: | 604 |
|---|
| Sequence: | MLARALLLCAVLALSHTANPCCSHPCQNRGVCMSVGFDQYKCDCTRTGFYGENCSTPEFL
TRIKLFLKPTPNTVHYILTHFKGFWNVVNNIPFLRNAIMSYVLTSRSHLIDSPPTYNADY
GYKSWEAFSNLSYYTRALPPVPDDCPTPLGVKGKKQLPDSNEIVEKLLLRRKFIPDPQGS
NMMFAFFAQHFTHQFFKTDHKRGPAFTNGLGHGVDLNHIYGETLARQRKLRLFKDGKMKY
QIIDGEMYPPTVKDTQAEMIYPPQVPEHLRFAVGQEVFGLVPGLMMYATIWLREHNRVCD
VLKQEHPEWGDEQLFQTSRLILIGETIKIVIEDYVQHLSGYHFKLKFDPELLFNKQFQYQ
NRIAAEFNTLYHWHPLLPDTFQIHDQKYNYQQFIYNNSILLEHGITQFVESFTRQIAGRV
AGGRNVPPAVQKVSQASIDQSRQMKYQSFNEYRKRFMLKPYESFEELTGEKEMSAELEAL
YGDIDAVELYPALLVEKPRPDAIFGETMVEVGAPFSLKGLMGNVICSPAYWKPSTFGGEV
GFQIINTASIQSLICNNVKGCPFTSFSVPDPELIKTVTINASSSRSGLDDINPTVLLKER
STEL
|
|
|
|---|
| BDBM50112547 |
|---|
| n/a |
|---|
| Name | BDBM50112547 |
|---|
| Synonyms: | 2-[1-(4-Chloro-benzoyl)-5-methoxy-2-methyl-1H-indol-3-yl]-N-(2-hydroxy-2-phenyl-ethyl)-acetamide | CHEMBL441769 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H25ClN2O4 |
|---|
| Mol. Mass. | 476.951 |
|---|
| SMILES | COc1ccc2n(C(=O)c3ccc(Cl)cc3)c(C)c(CC(=O)NC[C@@H](O)c3ccccc3)c2c1 |
|---|
| Structure | 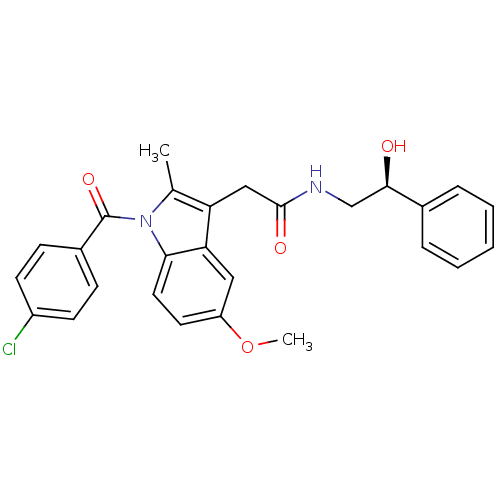
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kozak, KR; Prusakiewicz, JJ; Rowlinson, SW; Marnett, LJ Enantiospecific, selective cyclooxygenase-2 inhibitors. Bioorg Med Chem Lett12:1315-8 (2002) [PubMed]
Kozak, KR; Prusakiewicz, JJ; Rowlinson, SW; Marnett, LJ Enantiospecific, selective cyclooxygenase-2 inhibitors. Bioorg Med Chem Lett12:1315-8 (2002) [PubMed]