| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Neuromedin-K receptor |
|---|
| Ligand | BDBM50116728 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_209564 |
|---|
| Ki | 281.0±n/a nM |
|---|
| Citation |  Reichard, GA; Grice, CA; Shih, NY; Spitler, J; Majmundar, S; Wang, SD; Paliwal, S; Anthes, JC; Piwinski, JJ Preparation of oxime dual NK(1)/NK(2) antagonists with reduced NK(3) affinity. Bioorg Med Chem Lett12:2355-8 (2002) [PubMed] Reichard, GA; Grice, CA; Shih, NY; Spitler, J; Majmundar, S; Wang, SD; Paliwal, S; Anthes, JC; Piwinski, JJ Preparation of oxime dual NK(1)/NK(2) antagonists with reduced NK(3) affinity. Bioorg Med Chem Lett12:2355-8 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Neuromedin-K receptor |
|---|
| Name: | Neuromedin-K receptor |
|---|
| Synonyms: | NK-3 receptor | NK-3R | NK3R | NK3R_HUMAN | NKR | Neurokinin 3 receptor | Neurokinin B receptor | Neurokinin-3 (NK-3) | Neuromedin-3 receptor (NK-3R) | Neuromedin-3 receptor (NK3) | Neuromedin-K receptor | Neuromedin-K receptor (NK-3 receptor) | Neuromedin-K receptor (NK3) | Neuromedin-K receptor(NK3R) | TAC3R | TACR3 | Tachykinin receptor 3 | Tachykinin receptor 3 (NK3) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52221.96 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P29371 |
|---|
| Residue: | 465 |
|---|
| Sequence: | MATLPAAETWIDGGGGVGADAVNLTASLAAGAATGAVETGWLQLLDQAGNLSSSPSALGL
PVASPAPSQPWANLTNQFVQPSWRIALWSLAYGVVVAVAVLGNLIVIWIILAHKRMRTVT
NYFLVNLAFSDASMAAFNTLVNFIYALHSEWYFGANYCRFQNFFPITAVFASIYSMTAIA
VDRYMAIIDPLKPRLSATATKIVIGSIWILAFLLAFPQCLYSKTKVMPGRTLCFVQWPEG
PKQHFTYHIIVIILVYCFPLLIMGITYTIVGITLWGGEIPGDTCDKYHEQLKAKRKVVKM
MIIVVMTFAICWLPYHIYFILTAIYQQLNRWKYIQQVYLASFWLAMSSTMYNPIIYCCLN
KRFRAGFKRAFRWCPFIKVSSYDELELKTTRFHPNRQSSMYTVTRMESMTVVFDPNDADT
TRSSRKKRATPRDPSFNGCSRRNSKSASATSSFISSPYTSVDEYS
|
|
|
|---|
| BDBM50116728 |
|---|
| n/a |
|---|
| Name | BDBM50116728 |
|---|
| Synonyms: | 3,5-Dichloro-N-{(R)-3-(3,4-dichloro-phenyl)-5-[4-(3-ethylcarbamoylmethyl-2-oxo-2,3-dihydro-benzoimidazol-1-yl)-piperidin-1-yl]-2-[(Z)-methoxyimino]-pentyl}-N-ethyl-benzamide | CHEMBL430609 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C37H42Cl4N6O4 |
|---|
| Mol. Mass. | 776.579 |
|---|
| SMILES | CCNC(=O)Cn1c2ccccc2n(C2CCN(CC[C@@H](\C(CN(CC)C(=O)c3cc(Cl)cc(Cl)c3)=N\OC)c3ccc(Cl)c(Cl)c3)CC2)c1=O |
|---|
| Structure | 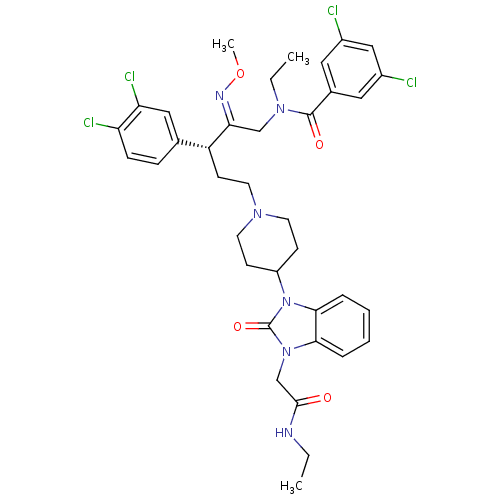
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Reichard, GA; Grice, CA; Shih, NY; Spitler, J; Majmundar, S; Wang, SD; Paliwal, S; Anthes, JC; Piwinski, JJ Preparation of oxime dual NK(1)/NK(2) antagonists with reduced NK(3) affinity. Bioorg Med Chem Lett12:2355-8 (2002) [PubMed]
Reichard, GA; Grice, CA; Shih, NY; Spitler, J; Majmundar, S; Wang, SD; Paliwal, S; Anthes, JC; Piwinski, JJ Preparation of oxime dual NK(1)/NK(2) antagonists with reduced NK(3) affinity. Bioorg Med Chem Lett12:2355-8 (2002) [PubMed]