| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM22988 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_68777 (CHEMBL686015) |
|---|
| Ki | >10000±n/a nM |
|---|
| Citation |  van der Stelt, M; van Kuik, JA; Bari, M; van Zadelhoff, G; Leeflang, BR; Veldink, GA; Finazzi-Agrò, A; Vliegenthart, JF; Maccarrone, M Oxygenated metabolites of anandamide and 2-arachidonoylglycerol: conformational analysis and interaction with cannabinoid receptors, membrane transporter, and fatty acid amide hydrolase. J Med Chem45:3709-20 (2002) [PubMed] van der Stelt, M; van Kuik, JA; Bari, M; van Zadelhoff, G; Leeflang, BR; Veldink, GA; Finazzi-Agrò, A; Vliegenthart, JF; Maccarrone, M Oxygenated metabolites of anandamide and 2-arachidonoylglycerol: conformational analysis and interaction with cannabinoid receptors, membrane transporter, and fatty acid amide hydrolase. J Med Chem45:3709-20 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH | FAAH1 | FAAH1_HUMAN | Fatty Acid Amide Hydrolase (FAAH) | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 63071.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00519 |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVQYELWAALPGASGVALACCFVAAAVALRWSGRRTARGAVVRARQRQRAGLENMDRAAQ
RFRLQNPDLDSEALLALPLPQLVQKLHSRELAPEAVLFTYVGKAWEVNKGTNCVTSYLAD
CETQLSQAPRQGLLYGVPVSLKECFTYKGQDSTLGLSLNEGVPAECDSVVVHVLKLQGAV
PFVHTNVPQSMFSYDCSNPLFGQTVNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSSFCGICGLKPTGNRLSKSGLKGCVYGQEAVRLSVGPMARDVESLALCLRALLCE
DMFRLDPTVPPLPFREEVYTSSQPLRVGYYETDNYTMPSPAMRRAVLETKQSLEAAGHTL
VPFLPSNIPHALETLSTGGLFSDGGHTFLQNFKGDFVDPCLGDLVSILKLPQWLKGLLAF
LVKPLLPRLSAFLSNMKSRSAGKLWELQHEIEVYRKTVIAQWRALDLDVVLTPMLAPALD
LNAPGRATGAVSYTMLYNCLDFPAGVVPVTTVTAEDEAQMEHYRGYFGDIWDKMLQKGMK
KSVGLPVAVQCVALPWQEELCLRFMREVERLMTPEKQSS
|
|
|
|---|
| BDBM22988 |
|---|
| n/a |
|---|
| Name | BDBM22988 |
|---|
| Synonyms: | (5Z,8Z,11Z,14Z)-N-(2-hydroxyethyl)icosa-5,8,11,14-tetraenamide | AEA | ANANDAMIDE | Arachidonoyl ethanolamide | Arachidonoyl ethanolamide (AEA) | Arachidonoylethanolamide (AEA) | Arachidonylethanolamide | CHEMBL15848 | [14C]Anandamide | [3H]Anandamide |
|---|
| Type | radiolabeled substrate |
|---|
| Emp. Form. | C22H37NO2 |
|---|
| Mol. Mass. | 347.5347 |
|---|
| SMILES | CCCCC\C=C/C\C=C/C\C=C/C\C=C/CCCC(=O)NCCO |
|---|
| Structure | 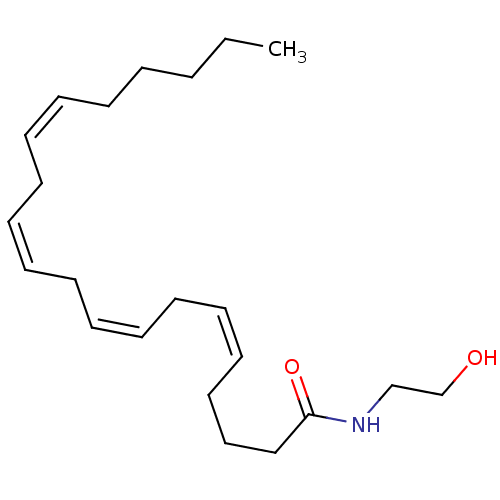
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 van der Stelt, M; van Kuik, JA; Bari, M; van Zadelhoff, G; Leeflang, BR; Veldink, GA; Finazzi-Agrò, A; Vliegenthart, JF; Maccarrone, M Oxygenated metabolites of anandamide and 2-arachidonoylglycerol: conformational analysis and interaction with cannabinoid receptors, membrane transporter, and fatty acid amide hydrolase. J Med Chem45:3709-20 (2002) [PubMed]
van der Stelt, M; van Kuik, JA; Bari, M; van Zadelhoff, G; Leeflang, BR; Veldink, GA; Finazzi-Agrò, A; Vliegenthart, JF; Maccarrone, M Oxygenated metabolites of anandamide and 2-arachidonoylglycerol: conformational analysis and interaction with cannabinoid receptors, membrane transporter, and fatty acid amide hydrolase. J Med Chem45:3709-20 (2002) [PubMed]