| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Ligand | BDBM50118594 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_212264 (CHEMBL816289) |
|---|
| IC50 | 27000±n/a nM |
|---|
| Citation |  Pirrung, MC; Tumey, LN; Raetz, CR; Jackman, JE; Snehalatha, K; McClerren, AL; Fierke, CA; Gantt, SL; Rusche, KM Inhibition of the antibacterial target UDP-(3-O-acyl)-N-acetylglucosamine deacetylase (LpxC): isoxazoline zinc amidase inhibitors bearing diverse metal binding groups. J Med Chem45:4359-70 (2002) [PubMed] Pirrung, MC; Tumey, LN; Raetz, CR; Jackman, JE; Snehalatha, K; McClerren, AL; Fierke, CA; Gantt, SL; Rusche, KM Inhibition of the antibacterial target UDP-(3-O-acyl)-N-acetylglucosamine deacetylase (LpxC): isoxazoline zinc amidase inhibitors bearing diverse metal binding groups. J Med Chem45:4359-70 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Name: | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Synonyms: | LPXC_ECOLI | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LpxC) | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LxpC) | UDP-3-O-acyl-GlcNAc deacetylase | asmB | envA | lpxC |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 33952.00 |
|---|
| Organism: | Escherichia coli |
|---|
| Description: | P0A725 |
|---|
| Residue: | 305 |
|---|
| Sequence: | MIKQRTLKRIVQATGVGLHTGKKVTLTLRPAPANTGVIYRRTDLNPPVDFPADAKSVRDT
MLCTCLVNEHDVRISTVEHLNAALAGLGIDNIVIEVNAPEIPIMDGSAAPFVYLLLDAGI
DELNCAKKFVRIKETVRVEDGDKWAEFKPYNGFSLDFTIDFNHPAIDSSNQRYAMNFSAD
AFMRQISRARTFGFMRDIEYLQSRGLCLGGSFDCAIVVDDYRVLNEDGLRFEDEFVRHKM
LDAIGDLFMCGHNIIGAFTAYKSGHALNNKLLQAVLAKQEAWEYVTFQDDAELPLAFKAP
SAVLA
|
|
|
|---|
| BDBM50118594 |
|---|
| n/a |
|---|
| Name | BDBM50118594 |
|---|
| Synonyms: | (R/S)-2-Mercapto-1-[3-(4-methoxy-phenyl)-4,5-dihydro-isoxazol-5-yl]-ethanone | CHEMBL134636 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H13NO3S |
|---|
| Mol. Mass. | 251.302 |
|---|
| SMILES | COc1ccc(cc1)C1=NOC(C1)C(=O)CS |t:9| |
|---|
| Structure | 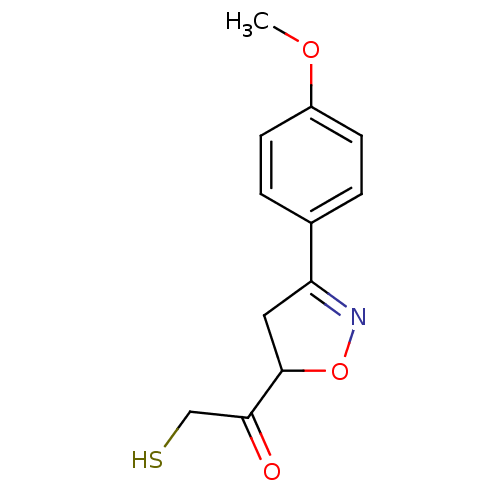
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Pirrung, MC; Tumey, LN; Raetz, CR; Jackman, JE; Snehalatha, K; McClerren, AL; Fierke, CA; Gantt, SL; Rusche, KM Inhibition of the antibacterial target UDP-(3-O-acyl)-N-acetylglucosamine deacetylase (LpxC): isoxazoline zinc amidase inhibitors bearing diverse metal binding groups. J Med Chem45:4359-70 (2002) [PubMed]
Pirrung, MC; Tumey, LN; Raetz, CR; Jackman, JE; Snehalatha, K; McClerren, AL; Fierke, CA; Gantt, SL; Rusche, KM Inhibition of the antibacterial target UDP-(3-O-acyl)-N-acetylglucosamine deacetylase (LpxC): isoxazoline zinc amidase inhibitors bearing diverse metal binding groups. J Med Chem45:4359-70 (2002) [PubMed]