

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | ||
| Ligand | BDBM50561541 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2075115 (CHEMBL4730649) | ||
| IC50 | 9400±n/a nM | ||
| Citation |  Vadukoot, AK; Sharma, S; Aretz, CD; Kumar, S; Gautam, N; Alnouti, Y; Aldrich, AL; Heim, CE; Kielian, T; Hopkins, CR Synthesis and SAR Studies of 1 ACS Med Chem Lett11:1848-1854 (2020) [PubMed] Article Vadukoot, AK; Sharma, S; Aretz, CD; Kumar, S; Gautam, N; Alnouti, Y; Aldrich, AL; Heim, CE; Kielian, T; Hopkins, CR Synthesis and SAR Studies of 1 ACS Med Chem Lett11:1848-1854 (2020) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| cAMP-specific 3',5'-cyclic phosphodiesterase 4B | |||
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | ||
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE4 | Isoform PDE4B1 | PDE32 | PDE4B | PDE4B1 | PDE4B_HUMAN | Phosphodiesterase 4B | Phosphodiesterase 4B (PDE4B) | Phosphodiesterase 4B (PDE4B1) | Phosphodiesterase Type 4 (PDE4B) | ||
| Type: | Protein | ||
| Mol. Mass.: | 83318.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q07343 | ||
| Residue: | 736 | ||
| Sequence: |
| ||
| BDBM50561541 | |||
| n/a | |||
| Name | BDBM50561541 | ||
| Synonyms: | CHEMBL4764438 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C19H17ClFN3O | ||
| Mol. Mass. | 357.809 | ||
| SMILES | Cc1ccc(cc1Cl)-n1c(cc2cccnc12)C(=O)NC1CC(F)C1 |(60.04,-30.68,;59.56,-32.15,;58.05,-32.48,;57.58,-33.94,;58.61,-35.08,;60.12,-34.76,;60.59,-33.29,;62.1,-32.97,;58.14,-36.54,;59.04,-37.79,;58.14,-39.03,;56.67,-38.56,;55.34,-39.34,;54,-38.57,;54.01,-37.02,;55.33,-36.25,;56.67,-37.01,;60.58,-37.78,;61.35,-36.45,;61.35,-39.12,;62.89,-39.12,;63.98,-40.21,;65.07,-39.12,;66.61,-39.12,;63.98,-38.03,)| | ||
| Structure | 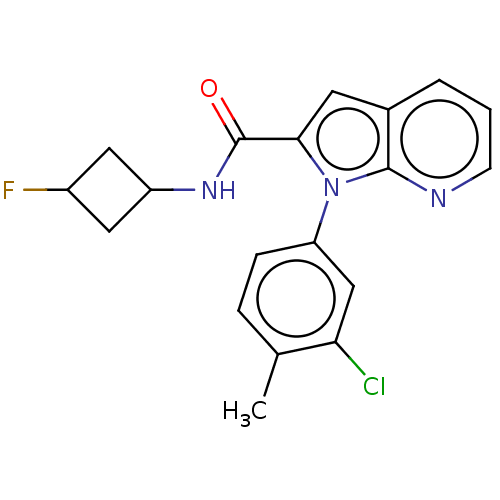
| ||