

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Nuclear factor NF-kappa-B p105 subunit | ||
| Ligand | BDBM50561592 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2075371 (CHEMBL4730905) | ||
| Kd | 216±n/a nM | ||
| Citation |  Annand, JR; Henderson, AR; Cole, KS; Maurais, AJ; Becerra, J; Liu, Y; Weerapana, E; Koehler, AN; Mapp, AK; Schindler, CS Gibberellin JRA-003: A Selective Inhibitor of Nuclear Translocation of IKK?. ACS Med Chem Lett11:1913-1918 (2020) [PubMed] Article Annand, JR; Henderson, AR; Cole, KS; Maurais, AJ; Becerra, J; Liu, Y; Weerapana, E; Koehler, AN; Mapp, AK; Schindler, CS Gibberellin JRA-003: A Selective Inhibitor of Nuclear Translocation of IKK?. ACS Med Chem Lett11:1913-1918 (2020) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Nuclear factor NF-kappa-B p105 subunit | |||
| Name: | Nuclear factor NF-kappa-B p105 subunit | ||
| Synonyms: | NFKB1 | NFKB1_HUMAN | Nuclear factor NF-kappa-B (NF-kB) | Nuclear factor NF-kappa-B complex | Nuclear factor NF-kappa-B p105 subunit | ||
| Type: | Protein | ||
| Mol. Mass.: | 105329.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P19838 | ||
| Residue: | 968 | ||
| Sequence: |
| ||
| BDBM50561592 | |||
| n/a | |||
| Name | BDBM50561592 | ||
| Synonyms: | (+)-Gibberellic Acid | CHEBI:28833 | GIBBERELLIN A3 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C19H22O6 | ||
| Mol. Mass. | 346.3744 | ||
| SMILES | [H][C@@]12CC[C@]3(O)C[C@]1(CC3=C)[C@@H](C(O)=O)[C@]1([H])[C@@]3(C)[C@@H](O)C=C[C@@]21OC3=O |r,wU:11.13,7.8,19.21,1.0,15.17,wD:4.4,23.27,17.19,c:23,TLB:20:19:15:24.25,THB:11:15:22.21.19:24.25,26:25:15:22.21.19,10:9:6:3.2.1,(.84,2.43,;.75,.58,;2.26,1.18,;3.44,.1,;3.11,-1.46,;4.56,-1.96,;1.77,-1.17,;.46,-1.03,;.82,-2.65,;2.42,-2.9,;3.13,-4.25,;-1.09,-1.15,;-1.86,-2.47,;-1.11,-3.8,;-3.39,-2.46,;-1.75,.24,;.07,.57,;-1.64,1.56,;-2.65,.41,;-2.61,2.98,;-4.12,3.15,;-1.48,4.02,;-1.62,2.79,;-.63,1.28,;1,1.21,;-.22,1.78,;0,3.29,)| | ||
| Structure | 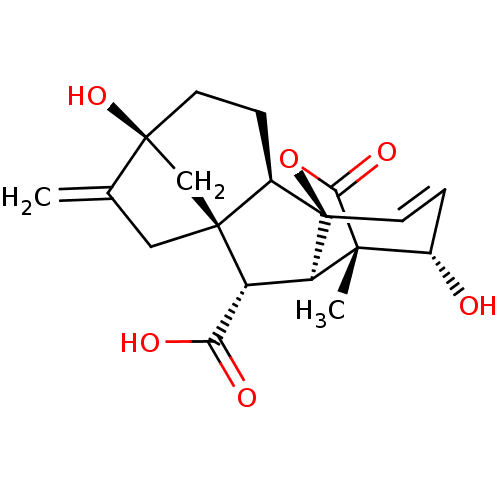
| ||